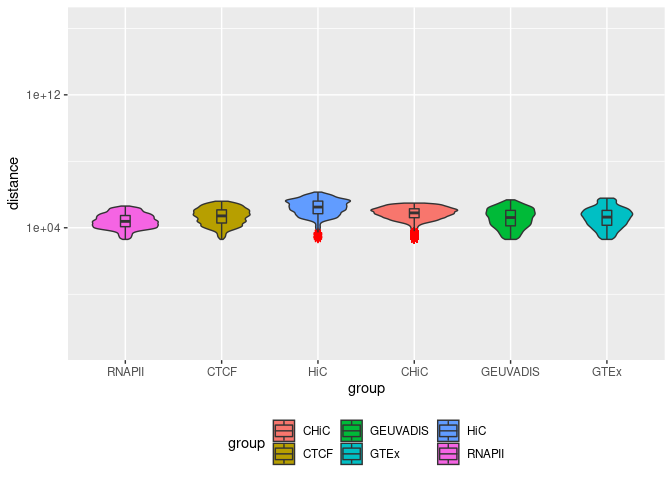
Summary statistics on BENGI GM12878 benchmarks
library(corrplot)
library(ggplot2)
Data importation
rm(list = ls())
nb_benchmarks <- 6
# Personal
path_to_benchmarks <- "/home/hoellinger/Documents/INSERM/BENGI/Benchmark/All-Pairs.Natural-Ratio/"
# Inserm path_to_benchmarks <-
# '/home/thoellinger/Documents/BENGI/Benchmark/All-Pairs.Natural-Ratio/'
# All pairs, fixed ratio, GM12878 ben1 <- 'GM12878.CHiC-Benchmark.v4.new.bedpe'
# ben2 <- 'GM12878.CTCF-ChIAPET-Benchmark.v4.new.bedpe' ben3 <-
# 'GM12878.GEUVADIS-Benchmark.v4.new.bedpe' ben4 <-
# 'GM12878.GTEx-Benchmark.v4.new.bedpe' ben5 <-
# 'GM12878.HiC-Benchmark.v4.new.bedpe' ben6 <-
# 'GM12878.RNAPII-ChIAPET-Benchmark.v4.new.bedpe' names <- c('CHiC', 'CTCF',
# 'GEUVADIS', 'GTEx', 'HiC', 'RNAPII') All pairs, natural ratio, GM12878
ben1 <- "GM12878.CHiC-Benchmark.v3.new.bedpe"
ben2 <- "GM12878.CTCF-ChIAPET-Benchmark.v3.new.bedpe"
ben3 <- "GM12878.GEUVADIS-Benchmark.v3.new.bedpe"
ben4 <- "GM12878.GTEx-Benchmark.v3.new.bedpe"
ben5 <- "GM12878.HiC-Benchmark.v3.new.bedpe"
ben6 <- "GM12878.RNAPII-ChIAPET-Benchmark.v3.new.bedpe"
names <- c("CHiC", "CTCF", "GEUVADIS", "GTEx", "HiC", "RNAPII")
list_of_files <- list(paste(path_to_benchmarks, ben1, sep = ""), paste(path_to_benchmarks,
ben2, sep = ""), paste(path_to_benchmarks, ben3, sep = ""), paste(path_to_benchmarks,
ben4, sep = ""), paste(path_to_benchmarks, ben5, sep = ""), paste(path_to_benchmarks,
ben6, sep = ""))
col_names <- c("chrom1", "start1", "end1", "chrom2", "start2", "end2", "name", "interaction",
"strand1", "strand2", "TSSs")
to_factor_benchmarks <- c(1, 4, 7, 8, 9, 10)
benchmarks = lapply(list_of_files, function(file) {
as.data.frame(read.table(file, sep = "\t"))
})
benchmarks = sapply(benchmarks, simplify = FALSE, function(Df) {
Df[to_factor_benchmarks] = lapply(Df[to_factor_benchmarks], factor)
names(Df) <- col_names
return(Df)
})
names(benchmarks) <- names
Overlap coefficient
positives <- list(nb_benchmarks)
positives <- sapply(benchmarks, simplify = FALSE, function(Df) {
return(Df$interaction == "1")
})
nb_positives <- vector(length = nb_benchmarks)
for (i in 1:nb_benchmarks) {
nb_positives[i] <- sum(positives[[i]])
}
length_benchmarks <- vector(length = nb_benchmarks) #seems it is not really useful
for (i in 1:nb_benchmarks) {
length_benchmarks[i] <- nrow(benchmarks[[i]])
}
Overlap <- as.matrix(diag(nb_benchmarks))
rownames(Overlap) <- names
colnames(Overlap) <- names
if (nb_benchmarks > 1) {
for (i in 2:nb_benchmarks) {
for (j in 1:(i - 1)) {
Overlap[i, j] = length(intersect(benchmarks[[i]][positives[[i]], 7],
benchmarks[[j]][positives[[j]], 7]))/min(nb_positives[i], nb_positives[j])
}
}
Overlap[upper.tri(Overlap)] <- t(Overlap)[upper.tri(Overlap)]
}
# png(file='/home/hoellinger/Documents/INSERM/shared/notes_perso/docs/moore2020/expA_all_pairs_fixed_ratio.png',
# width=700, height=700)
corrplot(Overlap, method = "color", type = "full", is.corr = FALSE, diag = TRUE,
outline = TRUE, addCoef.col = "black", addCoefasPercent = FALSE, order = "hclust",
hclust.method = "ward.D2")
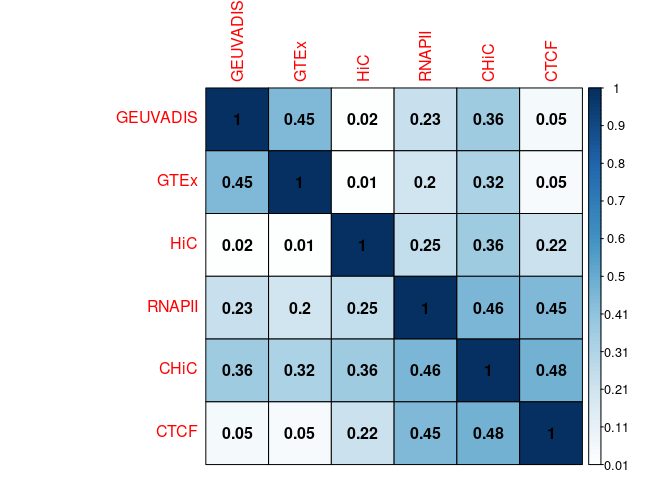
Distance distribution
benchmarks <- sapply(benchmarks, simplify = FALSE, USE.NAMES = TRUE, function(Df) {
Df[[11]] <- sapply(Df[[11]], simplify = FALSE, USE.NAMES = TRUE, function(x) {
x <- as.numeric(strsplit(x, ",")[[1]])
return(x)
})
return(Df)
})
# min of min distance
benchmarks <- sapply(benchmarks, simplify = FALSE, USE.NAMES = TRUE, function(Df) {
Df$distance <- mapply(function(x, y, z) min(min(abs(x - y)), min(abs(x - z))),
Df[[11]], Df[[5]], Df[[6]])
return(Df)
})
# Distances of positives + negatives Distances <- list() for(k in
# 1:nb_benchmarks){ Distances[[k]] <-
# data.frame(matrix(unlist(benchmarks[[k]]$distance), nrow=length_benchmarks[k],
# byrow=T), rep(names[k], length_benchmarks[k])) names(Distances[[k]]) <-
# c('distance', 'group') Distances[[k]]$group <- as.factor(Distances[[k]]$group)
# }
# Distances of positives
Distances <- list()
for (k in 1:nb_benchmarks) {
Distances[[k]] <- data.frame(matrix(unlist(benchmarks[[k]]$distance[positives[[k]]]),
nrow = nb_positives[k], byrow = T), rep(names[k], nb_positives[k]))
names(Distances[[k]]) <- c("distance", "group")
Distances[[k]]$group <- as.factor(Distances[[k]]$group)
}
Df_distances <- rbind(Distances[[1]], Distances[[2]], Distances[[3]], Distances[[4]],
Distances[[5]], Distances[[6]]) # should find a way to do this without having to manually write each element of the list
quantiles <- list()
for (k in 1:nb_benchmarks) {
quantiles[k] <- quantile(Distances[[k]]$distance, 0.95)
}
summary(Df_distances)
distance group
Min. : 2002 CHiC :88245
1st Qu.: 29068 CTCF : 7591
Median : 64854 GEUVADIS: 2073
Mean : 90392 GTEx : 1301
3rd Qu.: 127085 HiC : 3404
Max. :1402098 RNAPII :23699
print("0.95-quantiles:")
[1] "0.95-quantiles:"
print(names)
[1] "CHiC" "CTCF" "GEUVADIS" "GTEx" "HiC" "RNAPII"
t(quantiles)
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 245524.8 277808 282008 435494 890210.4 137229.7
# png(file='/home/hoellinger/Documents/INSERM/shared/notes_perso/docs/moore2020/distancesV1.png',
# width=700, height=700)
# png(file='/home/thoellinger/Documents/shared/notes_perso/docs/moore2020/distances_with_negatives_min_min.png',
# width=700, height=700) limits=c(1e-3,2e16)
ggplot(Df_distances, aes(x = group, y = distance, fill = group)) + geom_violin() +
scale_y_continuous(trans = "log10", limits = c(0.001, 2e+16), n.breaks = 3) +
geom_boxplot(width = 0.1, outlier.colour = "red", outlier.shape = 8) + theme(legend.position = "bottom") +
scale_x_discrete(limits = c(names[6], names[2], names[5], names[1], names[3],
names[4]))