Notice¶
The current page essentially corresponds to a markdown knitted from a Rmd document. It does not render very well because of html widgets not handled, but one should better refer to the correct rendering available here.
Libraries & Version¶
library(tidyverse)
library(visNetwork)
R version 4.0.3 (2020-10-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.1 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_GB.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_GB.UTF-8 LC_COLLATE=en_GB.UTF-8
[5] LC_MONETARY=en_GB.UTF-8 LC_MESSAGES=en_GB.UTF-8
[7] LC_PAPER=en_GB.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_GB.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] visNetwork_2.1.0 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7
[5] purrr_0.3.4 readr_2.0.2 tidyr_1.1.4 tibble_3.1.6
[9] ggplot2_3.3.5 tidyverse_1.3.1 knitr_1.36
loaded via a namespace (and not attached):
[1] tidyselect_1.1.1 xfun_0.28 bslib_0.3.1 haven_2.4.3
[5] colorspace_2.0-2 vctrs_0.3.8 generics_0.1.1 htmltools_0.5.2
[9] yaml_2.2.1 utf8_1.2.2 rlang_0.4.12 jquerylib_0.1.4
[13] pillar_1.6.4 withr_2.4.2 glue_1.5.0 DBI_1.1.1
[17] dbplyr_2.1.1 modelr_0.1.8 readxl_1.3.1 lifecycle_1.0.1
[21] munsell_0.5.0 gtable_0.3.0 cellranger_1.1.0 rvest_1.0.2
[25] htmlwidgets_1.5.4 evaluate_0.14 tzdb_0.2.0 fastmap_1.1.0
[29] fansi_0.5.0 broom_0.7.10 Rcpp_1.0.7 backports_1.3.0
[33] scales_1.1.1 formatR_1.11 jsonlite_1.7.2 fs_1.5.0
[37] hms_1.1.1 digest_0.6.28 stringi_1.7.5 grid_4.0.3
[41] cli_3.1.0 tools_4.0.3 magrittr_2.0.1 sass_0.4.0
[45] crayon_1.4.2 pkgconfig_2.0.3 ellipsis_0.3.2 xml2_1.3.2
[49] reprex_2.0.1 lubridate_1.8.0 rstudioapi_0.13 assertthat_0.2.1
[53] rmarkdown_2.11 httr_1.4.2 R6_2.5.1 compiler_4.0.3
Preliminary work¶
The documentation on visNetwork is available here.
Data importation¶
All the files imported here can be found on Genotoul, in (private access) /work2/project/regenet/workspace/thoellinger/shared/2022/promoter_capture_hic/ and (private access) /work2/project/regenet/results/phic/homo_sapiens/hg19/jung.ren.2019/LI11/nofilter/pp, or for some here (public access).
The full code itself is available here: /work2/project/regenet/workspace/thoellinger/shared/2022/promoter_capture_hic/ (as preliminary_analysis_v1.Rmd). Please do not modify directly this repository as it is backed up between multiple computers on a regular basis.
rm(list = ls())
# wd = 'data/'
wd = "/home/thoellinger/Documents/shared/2022/promoter_capture_hic/data/"
# wd =
# '/home/hoellinger/Documents/INSERM/shared/2022/promoter_capture_hic/data/'
egfile = paste(wd, "liver.all_putative_enhancers.overlapping_ccRE-ELS.sorted.bedpe",
sep = "")
merged_efile = paste(wd, "list_all_enhancers.overlapping_ccRE-ELS.bed", sep = "")
############# Enhancers #
me = as.data.frame(read.table(merged_efile, sep = "\t"))
############ E-G list #
eg = as.data.frame(read.table(egfile, header = F, col.names = c("chrom1", "start1",
"end1", "chrom2", "start2", "end2", "name", "score.contact", "strand1", "strand2",
"tissue", "gene.symbol", "original.distance", "gene.id"), sep = "\t"))
############### known genes #
gene_list = c("HFE", "TFR2", "HFE2", "HAMP", "SLC40A1", "BMP6", "TMPRSS6", "TFRC",
"SLC11A2", "CYBRD1", "NEO1", "CIAPIN1", "SLC39A14")
# genes known to be causally involved in hemochromatosis: 'HFE', 'TFR2',
# 'HFE2', 'HAMP', 'SLC40A1', 'BMP6' (the other genes are involved in iron
# metabolism regulation) genes co-expressed in the liver: HFE + TFR2 + HJV +
# HAMP + TMPRSS6 genes co-expressed in intestine: DCYTB + DMT1 + SLC40A1
Data preprocessing¶
Conversion to factors¶
# to_factor_cols = c('chrom1', 'chrom2', 'name', 'strand1', 'strand2',
# 'tissue', 'gene.symbol', 'gene.id')
to_factor_cols = c("chrom1", "chrom2", "name", "strand1", "strand2", "tissue")
eg[to_factor_cols] = lapply(eg[to_factor_cols], factor)
Exploration¶
Summary statistics on enhancer lists¶
length(me[, 1])
[1] 25600
Warning: we shall pay attention to the fact that those 25,600 putative enhancers correspond to those, among the 31,749 initial putative regulatory elements in the CHiC data, that intersect at least one ccRE-ELS. We filtered out the other elements / the E-G pairs involving such elements. We did so because they are pretty large (see next cell), so it is very unlikely for such large element to be an enhancer but not to intersect any ccRE-ELS.
me$length = abs(me$V3 - me$V2)
summary(me$length)
Min. 1st Qu. Median Mean 3rd Qu. Max.
1571 4386 5843 7324 8198 122914
E-G pairs¶
We extract the subsample of the E-G bedpe input list, where genes are contained in our list of genes involved either directly in hemochromatosis or in iron metabolism. In the variable name, "dist0" stands for "distance is 0 between the genes in eg_dist0 and the list of initial genes".
eg_dist0 = eg[eg$gene.symbol %in% gene_list, ]
length(eg_dist0$name)
[1] 49
length(unique(eg_dist0$gene.symbol))
[1] 12
print(as.character(unique(eg_dist0$gene.symbol)))
[1] "HFE2" "CYBRD1" "SLC40A1" "TFRC" "BMP6" "HFE"
[7] "TFR2" "SLC39A14" "SLC11A2" "NEO1" "CIAPIN1" "TMPRSS6"
print(gene_list)
[1] "HFE" "TFR2" "HFE2" "HAMP" "SLC40A1" "BMP6"
[7] "TMPRSS6" "TFRC" "SLC11A2" "CYBRD1" "NEO1" "CIAPIN1"
[13] "SLC39A14"
We have element-gene pairs in our data for 12 out of 13 initial genes.
Genes¶
Chromosomes where the genes are located:
HFE TFR2 HFE2 HAMP SLC40A1 BMP6 TMPRSS6 TFRC
"chr6" "chr7" "chr1" "HAMP" "chr2" "chr6" "chr22" "chr3"
SLC11A2 CYBRD1 NEO1 CIAPIN1 SLC39A14
"chr12" "chr2" "chr15" "chr16" "chr8"
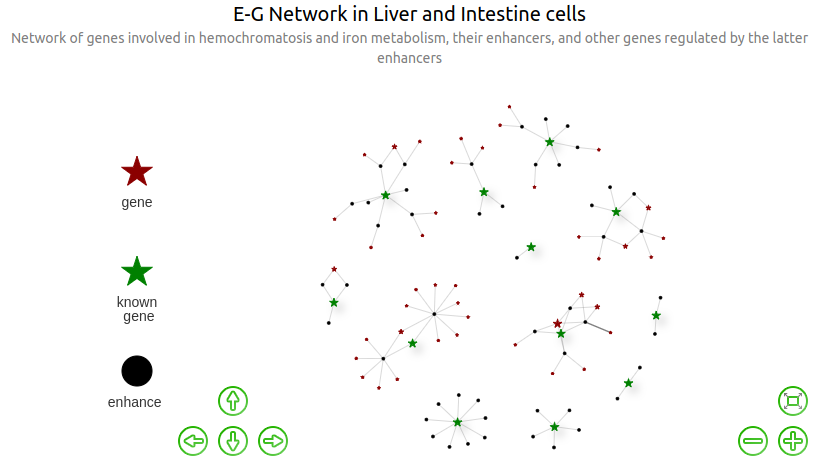
Networks¶
Note: in all subsequent graphs, size of nodes of type "gene" (and not "known_gene", for which the size is fixed) is proportional to the number of distinct enhancers regulating them.
Find all genes regulated by the initial enhancers¶
The "initial enhancers" are the enhancers involved in eg_dist0, ie all the enhancers regulating the initial genes in gene_list.
genes is the subset of gene_list for which we have data (12 out of 13 genes here), and enhancers is the list of enhancers regulating those initial known genes.
enhancers = unique(paste(eg_dist0$chrom1, ":", eg_dist0$start1, "-", eg_dist0$end1,
sep = ""))
genes = unique(eg_dist0$gene.symbol)
Now we compute the list of all genes regulated by enhancers in eg_dist0. Specifically, we extract, from the full E-G list eg, the list eg_dist1 containing only the enhancers-genes pairs for which the gene G is regulated by any of the enhancers regulating a gene in gene_list ("dist1" stands for "distance is at most 1 between the genes in eg_dist1 and the list of initial genes").
eg_enhancers_id = data.frame(source = paste(eg$chrom1, ":", eg$start1, "-", eg$end1,
sep = ""), eg[, -c(1, 2, 3)]) # same as eg but columns 1-3 have been concatenated to make unique enhancers id
eg_dist1 = eg_enhancers_id[eg_enhancers_id$source %in% paste(eg_dist0$chrom1, ":",
eg_dist0$start1, "-", eg_dist0$end1, sep = ""), ]
eg_dist1$from = lapply(eg_dist1$source, function(x) unique(as.character(eg_dist0[paste(eg_dist0$chrom1,
":", eg_dist0$start1, "-", eg_dist0$end1, sep = "") == x, "gene.symbol"])))
eg_dist1$from.id = lapply(eg_dist1$source, function(x) unique(as.character(eg_dist0[paste(eg_dist0$chrom1,
":", eg_dist0$start1, "-", eg_dist0$end1, sep = "") == x, "gene.id"])))
eg_dist1$score.contact.IE = left_join(data.frame(name = paste(eg_dist1$source, eg_dist1$from.id,
eg_dist1$from, sep = "::")), eg_enhancers_id, by = "name")$score.contact # max ABC score of the I-E pair (initialGene-Enhancer) corresponding to the E-G pair
genes_dist1 = unique(eg_dist1$gene.symbol)
genes_dist1 is the list of genes regulated by enhancers.
Compute "contact product", ie the product of the contact scores of: - the initial gene - enhancer pair (I-E) - the enhancer - gene pair (E-G)
eg_dist1$contact.product = eg_dist1$score.contact * eg_dist1$score.contact.IE
eg_dist1$contact.product = eg_dist1$contact.product/max(eg_dist1$contact.product)
print(min(eg_dist1$contact.product))
[1] 0.1613721
print(mean(eg_dist1$contact.product))
[1] 0.3154293
print(max(eg_dist1$contact.product))
[1] 1
print(quantile(eg_dist1$contact.product, c(0.1, 0.4, 0.5, 0.6, 0.8, 0.9)))
10% 40% 50% 60% 80% 90%
0.1853717 0.2340752 0.2457107 0.2883543 0.4243637 0.5197058
eg_dist1$contact.product.label = 1
eg_dist1[eg_dist1$contact.product >= median(eg_dist1$contact.product), ]$contact.product.label = 2
eg_dist1[eg_dist1$contact.product >= quantile(eg_dist1$contact.product, 0.9)[[1]],
]$contact.product.label = 3
table(eg_dist1$contact.product.label)
1 2 3
50 39 12
In the following cell we re-organize genes_dist1 into genes_dist1.more which is in a well-suited format for further "concatenation" with inferences made with other type of data (ABC or QTL -based).
genes_dist1.more = eg_dist1 %>%
group_by(gene.symbol) %>%
mutate(CHiC.sources = paste0(source, collapse = ",")) %>%
mutate(CHiC.count = length(str_split(CHiC.sources, ",")[[1]])) %>%
slice(which.max(contact.product.label)) %>%
ungroup() %>%
select(gene.symbol, gene.id, CHiC.sources, contact.product.label, CHiC.count,
from)
genes_dist1.more = subset(genes_dist1.more, !(genes_dist1.more$gene.symbol %in% genes))
genes_dist1.more$from = as.character(genes_dist1.more$from)
genes_dist1.more
# A tibble: 42 × 6
gene.symbol gene.id CHiC.sources contact.product… CHiC.count from
<chr> <chr> <chr> <dbl> <int> <chr>
1 ACTL6B ENSG00000077080.5 chr7:100151… 1 2 TFR2
2 ADPGK ENSG00000159322.13 chr15:73089… 3 2 NEO1
3 AGFG2 ENSG00000106351.8 chr7:100151… 1 1 TFR2
4 ANKRD34A ENSG00000181039.7 chr1:145508… 2 1 HFE2
5 ARL2BP ENSG00000102931.3 chr16:57256… 2 1 CIAP…
6 ATF1 ENSG00000123268.4 chr12:51316… 2 1 SLC1…
7 C1QTNF6 ENSG00000133466.9 chr22:37461… 2 2 TMPR…
8 C7orf61 ENSG00000185955.4 chr7:100151… 1 1 TFR2
9 CCDC135 ENSG00000159625.10 chr16:57491… 3 2 CIAP…
10 CCL22 ENSG00000102962.4 chr16:57428… 2 1 CIAP…
# … with 32 more rows
We re-arrange eg_dist1 as an edge list edges_list_dist1, which will be more suitable to later construct the edges list for visualization as a graph.
edges_list_dist1 = data.frame(source = eg_dist1$source, target = eg_dist1$gene.symbol,
distance_kb = floor(eg_dist1$original.distance/1000), inv_dist = 1/(eg_dist1$original.distance +
1), rescaled_log_inv_dist = 1 - min(log(1/(eg_dist1$original.distance + 1))) +
log(1/(eg_dist1$original.distance + 1)))
edges_list_dist1
Now we can compute the list nodes_dist1 of (colored) nodes required for our graphs. There are 3 types of nodes: enhancer, known_gene and (unknown) gene.
nodes_dist1 = full_join(data.frame(label = unique(eg_dist1$source), group = "enhancer"),
data.frame(label = unique(eg_dist1$gene.symbol), group = "gene")) %>%
rowid_to_column("id")
nodes_dist1[nodes_dist1$label %in% genes, ]$group = "known_gene"
nodes_dist1 = unique(nodes_dist1)
table(nodes_dist1$group)
enhancer gene known_gene
49 42 12
In the list of edges of the graph, edges_dist1, the sample column indicates in which family of tissues (liver, intestine or both) each E-G pair has been found.
We add to nodes_dist1 a column d_in for plotting purpose: it contains 1 for each node of type enhancer, the number of incoming enhancers for each node of type gene, and the max of the latter for each node of type known_gene.
Edge weight based on distance¶
Width proportional to distance. For each enhancer-gene pair $E$-$G$, the distance indicated in the eg dataframe as original_distance.mean is given in in base pairs. Here, distance_kb is the very same quantity but expressed in kb.
Width proportional to inverse distance:
Width proportional to rescaled and translated log inverse distance:
Results¶
The list of the initial genes + all genes regulated by enhancers regulating the 12 initial genes (12 found among 13), can be found here: /work2/project/regenet/workspace/thoellinger/shared/automne_2021/promoter_capture_hic/results/new_genes_v1.list.
# write.table(genes_dist1, file='results/new_genes_v1.list', quote=FALSE,
# sep='\t', row.names=F, col.names=F)
The list all genes regulated by enhancers regulating the 12 initial genes, with useful info, can be found here: /work2/project/regenet/workspace/thoellinger/shared/2022/promoter_capture_hic/results/new_genes_chic_v2_more_info.list.
# write.table(genes_dist1.more,
# file='results/new_genes_chic_v2_more_info.list', quote=FALSE, sep='\t',
# row.names=F, col.names=T)
For each one of the 12 initial genes, the list of genes regulated by enhancers regulating that initial gene, can be found here:
# for (gene in genes){ current_gene_dist1 =
# unique(eg_dist1[eg_dist1$from==gene,'gene.symbol'])
# write.table(current_gene_dist1, file=paste('results/separate/',gene,'.list',
# sep=''), quote=FALSE, sep='\t', row.names=F, col.names=F) }
The list of the enhancers regulating the 12 initial genes, can be found here: /work2/project/regenet/workspace/thoellinger/shared/2022/promoter_capture_hic/results/enhancers.list
# write.table(enhancers, file='results/enhancers.list', quote=FALSE, sep='\t',
# row.names=F, col.names=F)
The list of E-G pairs involving the 12 initial genes, can be found here: /work2/project/regenet/workspace/thoellinger/shared/2022/promoter_capture_hic/results/eg_dist0.bedpe
# write.table(eg_dist0, file='results/eg_dist0.bedpe', quote=FALSE, sep='\t',
# row.names=F, col.names=F)
The list of the enhancers regulating the 54 initial+new genes, can be found here: /work2/project/regenet/workspace/thoellinger/shared/2022/promoter_capture_hic/results/enhancers.new_genes.list
# eg_dist2 = eg[eg$gene.symbol %in% genes_dist1,] enhancers.new_genes =
# unique(paste(eg_dist2$chrom1, ':', eg_dist2$start1, '-', eg_dist2$end1,
# sep='')) write.table(enhancers.new_genes,
# file='results/enhancers.new_genes.list', quote=FALSE, sep='\t', row.names=F,
# col.names=F)
The list of E-G pairs involving the 54 initial+new genes, can be found here: /work2/project/regenet/workspace/thoellinger/shared/2022/promoter_capture_hic/results/eg_dist2.bedpe
# write.table(eg_dist2, file='results/eg_dist2.bedpe', quote=FALSE, sep='\t',
# row.names=F, col.names=F)
Appendix¶
Tables¶
Table of the initial ABC-predicted E-G pairs involving the initial genes¶
options(max.print = 1000)
eg_dist0
chrom1 start1 end1 chrom2 start2 end2
1602 chr1 144995955 144999800 chr1 145413094 145417545
1613 chr1 145382770 145387982 chr1 145413094 145417545
1620 chr1 145445045 145450003 chr1 145413094 145417545
1622 chr1 145471844 145478745 chr1 145413094 145417545
1626 chr1 145508505 145515028 chr1 145413094 145417545
1627 chr1 145519579 145527792 chr1 145413094 145417545
3982 chr2 171868841 171871980 chr2 172378756 172414643
3983 chr2 172052913 172056719 chr2 172378756 172414643
3985 chr2 172244003 172248890 chr2 172378756 172414643
3986 chr2 172359194 172365051 chr2 172378756 172414643
3987 chr2 172396460 172401984 chr2 172378756 172414643
4074 chr2 190325771 190329372 chr2 190425304 190448484
4075 chr2 190329373 190334233 chr2 190425304 190448484
4076 chr2 190360995 190365156 chr2 190425304 190448484
4077 chr2 190390528 190393955 chr2 190425304 190448484
4078 chr2 190410593 190416234 chr2 190425304 190448484
4079 chr2 190416235 190421209 chr2 190425304 190448484
4080 chr2 190421210 190425029 chr2 190425304 190448484
4081 chr2 190475143 190478818 chr2 190425304 190448484
4085 chr2 190496245 190499388 chr2 190425304 190448484
6360 chr3 195829327 195839503 chr3 195754053 195809060
6361 chr3 195904445 195910895 chr3 195754053 195809060
9222 chr6 7757920 7762092 chr6 7727029 7881655
9223 chr6 8000333 8004738 chr6 7727029 7881655
9356 chr6 26065333 26073782 chr6 26087508 26098571
12009 chr7 100151130 100158146 chr7 100218038 100240402
12040 chr7 100479857 100484394 chr7 100218038 100240402
12830 chr8 22136614 22143599 chr8 22224761 22291642
12831 chr8 22143600 22148473 chr8 22224761 22291642
12835 chr8 22148474 22155405 chr8 22224761 22291642
20296 chr12 50495719 50503727 chr12 51373183 51422349
20338 chr12 51184932 51188867 chr12 51373183 51422349
20341 chr12 51316042 51321932 chr12 51373183 51422349
20362 chr12 51540821 51574422 chr12 51373183 51422349
20367 chr12 51637790 51642516 chr12 51373183 51422349
20369 chr12 51652001 51656616 chr12 51373183 51422349
20373 chr12 51670781 51676690 chr12 51373183 51422349
25072 chr15 73089434 73099467 chr15 73344050 73597547
25074 chr15 73177101 73180543 chr15 73344050 73597547
25078 chr15 73372310 73380150 chr15 73344050 73597547
26940 chr16 57256998 57262124 chr16 57462080 57481440
26952 chr16 57428584 57434799 chr16 57462080 57481440
26967 chr16 57491859 57510132 chr16 57462080 57481440
26985 chr16 57662892 57669586 chr16 57462080 57481440
38401 chr22 36318565 36323139 chr22 37461475 37505603
38476 chr22 37451969 37461031 chr22 37461475 37505603
38481 chr22 37461032 37471647 chr22 37461475 37505603
38484 chr22 37533344 37544456 chr22 37461475 37505603
38485 chr22 37550713 37556000 chr22 37461475 37505603
name score.contact
1602 chr1:144995955-144999800::ENSG00000168509.13::HFE2 2.691613
1613 chr1:145382770-145387982::ENSG00000168509.13::HFE2 3.500083
1620 chr1:145445045-145450003::ENSG00000168509.13::HFE2 2.747677
1622 chr1:145471844-145478745::ENSG00000168509.13::HFE2 2.173732
1626 chr1:145508505-145515028::ENSG00000168509.13::HFE2 3.404033
1627 chr1:145519579-145527792::ENSG00000168509.13::HFE2 3.329819
3982 chr2:171868841-171871980::ENSG00000071967.7::CYBRD1 3.161168
3983 chr2:172052913-172056719::ENSG00000071967.7::CYBRD1 2.180688
3985 chr2:172244003-172248890::ENSG00000071967.7::CYBRD1 2.113738
3986 chr2:172359194-172365051::ENSG00000071967.7::CYBRD1 2.961889
3987 chr2:172396460-172401984::ENSG00000071967.7::CYBRD1 2.013556
4074 chr2:190325771-190329372::ENSG00000138449.6::SLC40A1 4.556188
4075 chr2:190329373-190334233::ENSG00000138449.6::SLC40A1 2.672282
4076 chr2:190360995-190365156::ENSG00000138449.6::SLC40A1 3.338258
4077 chr2:190390528-190393955::ENSG00000138449.6::SLC40A1 2.310444
4078 chr2:190410593-190416234::ENSG00000138449.6::SLC40A1 2.833319
4079 chr2:190416235-190421209::ENSG00000138449.6::SLC40A1 2.538705
4080 chr2:190421210-190425029::ENSG00000138449.6::SLC40A1 2.206945
4081 chr2:190475143-190478818::ENSG00000138449.6::SLC40A1 2.561220
4085 chr2:190496245-190499388::ENSG00000138449.6::SLC40A1 2.306987
6360 chr3:195829327-195839503::ENSG00000072274.8::TFRC 2.880979
6361 chr3:195904445-195910895::ENSG00000072274.8::TFRC 2.461830
9222 chr6:7757920-7762092::ENSG00000153162.8::BMP6 3.182610
9223 chr6:8000333-8004738::ENSG00000153162.8::BMP6 2.373608
9356 chr6:26065333-26073782::ENSG00000010704.14::HFE 2.104796
12009 chr7:100151130-100158146::ENSG00000106327.8::TFR2 2.202777
12040 chr7:100479857-100484394::ENSG00000106327.8::TFR2 2.085438
12830 chr8:22136614-22143599::ENSG00000104635.9::SLC39A14 2.941420
12831 chr8:22143600-22148473::ENSG00000104635.9::SLC39A14 2.234129
12835 chr8:22148474-22155405::ENSG00000104635.9::SLC39A14 2.240686
20296 chr12:50495719-50503727::ENSG00000110911.10::SLC11A2 2.207329
20338 chr12:51184932-51188867::ENSG00000110911.10::SLC11A2 2.739063
20341 chr12:51316042-51321932::ENSG00000110911.10::SLC11A2 2.933383
20362 chr12:51540821-51574422::ENSG00000110911.10::SLC11A2 2.623970
20367 chr12:51637790-51642516::ENSG00000110911.10::SLC11A2 2.607919
20369 chr12:51652001-51656616::ENSG00000110911.10::SLC11A2 2.923081
20373 chr12:51670781-51676690::ENSG00000110911.10::SLC11A2 2.106838
25072 chr15:73089434-73099467::ENSG00000067141.12::NEO1 3.560153
25074 chr15:73177101-73180543::ENSG00000067141.12::NEO1 2.328833
25078 chr15:73372310-73380150::ENSG00000067141.12::NEO1 3.416054
26940 chr16:57256998-57262124::ENSG00000005194.10::CIAPIN1 2.036134
26952 chr16:57428584-57434799::ENSG00000005194.10::CIAPIN1 5.012442
26967 chr16:57491859-57510132::ENSG00000005194.10::CIAPIN1 2.158098
26985 chr16:57662892-57669586::ENSG00000005194.10::CIAPIN1 3.803306
38401 chr22:36318565-36323139::ENSG00000187045.12::TMPRSS6 2.429089
38476 chr22:37451969-37461031::ENSG00000187045.12::TMPRSS6 2.256193
38481 chr22:37461032-37471647::ENSG00000187045.12::TMPRSS6 3.763882
38484 chr22:37533344-37544456::ENSG00000187045.12::TMPRSS6 2.021822
38485 chr22:37550713-37556000::ENSG00000187045.12::TMPRSS6 2.426097
strand1 strand2 tissue gene.symbol original.distance gene.id
1602 . . liver HFE2 411368 ENSG00000168509.13
1613 . . liver HFE2 23186 ENSG00000168509.13
1620 . . liver HFE2 25241 ENSG00000168509.13
1622 . . liver HFE2 52040 ENSG00000168509.13
1626 . . liver HFE2 88701 ENSG00000168509.13
1627 . . liver HFE2 99775 ENSG00000168509.13
3982 . . liver CYBRD1 501217 ENSG00000071967.7
3983 . . liver CYBRD1 316478 ENSG00000071967.7
3985 . . liver CYBRD1 124307 ENSG00000071967.7
3986 . . liver CYBRD1 8146 ENSG00000071967.7
3987 . . liver CYBRD1 14161 ENSG00000071967.7
4074 . . liver SLC40A1 104295 ENSG00000138449.6
4075 . . liver SLC40A1 99434 ENSG00000138449.6
4076 . . liver SLC40A1 68511 ENSG00000138449.6
4077 . . liver SLC40A1 39712 ENSG00000138449.6
4078 . . liver SLC40A1 17433 ENSG00000138449.6
4079 . . liver SLC40A1 12458 ENSG00000138449.6
4080 . . liver SLC40A1 8638 ENSG00000138449.6
4081 . . liver SLC40A1 24286 ENSG00000138449.6
4085 . . liver SLC40A1 45388 ENSG00000138449.6
6360 . . liver TFRC 10918 ENSG00000072274.8
6361 . . liver TFRC 86036 ENSG00000072274.8
9222 . . liver BMP6 23130 ENSG00000153162.8
9223 . . liver BMP6 265543 ENSG00000153162.8
9356 . . liver HFE 13117 ENSG00000010704.14
12009 . . liver TFR2 71635 ENSG00000106327.8
12040 . . liver TFR2 224731 ENSG00000106327.8
12830 . . liver SLC39A14 79896 ENSG00000104635.9
12831 . . liver SLC39A14 75022 ENSG00000104635.9
12835 . . liver SLC39A14 68090 ENSG00000104635.9
20296 . . liver SLC11A2 913646 ENSG00000110911.10
20338 . . liver SLC11A2 228506 ENSG00000110911.10
20341 . . liver SLC11A2 95441 ENSG00000110911.10
20362 . . liver SLC11A2 106378 ENSG00000110911.10
20367 . . liver SLC11A2 203347 ENSG00000110911.10
20369 . . liver SLC11A2 217558 ENSG00000110911.10
20373 . . liver SLC11A2 236338 ENSG00000110911.10
25072 . . liver NEO1 242730 ENSG00000067141.12
25074 . . liver NEO1 161654 ENSG00000067141.12
25078 . . liver NEO1 20925 ENSG00000067141.12
26940 . . liver CIAPIN1 214245 ENSG00000005194.10
26952 . . liver CIAPIN1 41570 ENSG00000005194.10
26967 . . liver CIAPIN1 7286 ENSG00000005194.10
26985 . . liver CIAPIN1 178319 ENSG00000005194.10
38401 . . liver TMPRSS6 1156734 ENSG00000187045.12
38476 . . liver TMPRSS6 18842 ENSG00000187045.12
38481 . . liver TMPRSS6 8226 ENSG00000187045.12
38484 . . liver TMPRSS6 17904 ENSG00000187045.12
38485 . . liver TMPRSS6 35273 ENSG00000187045.12
options(max.print = 75)
List of the infered new genes¶
options(max.print = 1000)
genes_dist1
[1] "HFE2" "PDE4DIP" "NUDT17" "POLR3GL" "ANKRD34A" "CYBRD1"
[7] "SLC40A1" "TFRC" "BMP6" "HFE" "C7orf61" "TSC22D4"
[13] "ACTL6B" "AGFG2" "TFR2" "NYAP1" "PCOLCE" "FBXO24"
[19] "CLDN15" "SAP25" "TRIM56" "FIS1" "EPO" "LRCH4"
[25] "ZNHIT1" "SLC39A14" "POLR3D" "PHYHIP" "XPO7" "SLC11A2"
[31] "DAZAP2" "CERS5" "METTL7A" "CSRNP2" "ATF1" "NR4A1"
[37] "CELA1" "NEO1" "ADPGK" "ARL2BP" "CIAPIN1" "COQ9"
[43] "CCL22" "CX3CL1" "KATNB1" "CCDC135" "DOK4" "TMPRSS6"
[49] "KCTD17" "C1QTNF6" "RAC2" "IL2RB" "MPST" "TST"
length(genes_dist1)
[1] 54
options(max.print = 75)