Notice¶
The current page essentially corresponds to a markdown knitted from a Rmd document. It does not render very well because of html widgets not handled, so one should better refer to the correct rendering available in preliminary_analysis_v9.html.
Libraries & Version¶
library(tidyverse)
library(visNetwork)
library(data.table)
R version 4.0.3 (2020-10-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.1 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_GB.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_GB.UTF-8 LC_COLLATE=en_GB.UTF-8
[5] LC_MONETARY=en_GB.UTF-8 LC_MESSAGES=en_GB.UTF-8
[7] LC_PAPER=en_GB.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_GB.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] data.table_1.14.2 visNetwork_2.1.0 forcats_0.5.1 stringr_1.4.0
[5] dplyr_1.0.7 purrr_0.3.4 readr_2.0.2 tidyr_1.1.4
[9] tibble_3.1.6 ggplot2_3.3.5 tidyverse_1.3.1 knitr_1.36
loaded via a namespace (and not attached):
[1] tidyselect_1.1.1 xfun_0.28 bslib_0.3.1 haven_2.4.3
[5] colorspace_2.0-2 vctrs_0.3.8 generics_0.1.1 htmltools_0.5.2
[9] yaml_2.2.1 utf8_1.2.2 rlang_0.4.12 jquerylib_0.1.4
[13] pillar_1.6.4 withr_2.4.2 glue_1.5.0 DBI_1.1.1
[17] dbplyr_2.1.1 modelr_0.1.8 readxl_1.3.1 lifecycle_1.0.1
[21] munsell_0.5.0 gtable_0.3.0 cellranger_1.1.0 rvest_1.0.2
[25] htmlwidgets_1.5.4 evaluate_0.14 tzdb_0.2.0 fastmap_1.1.0
[29] fansi_0.5.0 broom_0.7.10 Rcpp_1.0.7 backports_1.3.0
[33] scales_1.1.1 formatR_1.11 jsonlite_1.7.2 fs_1.5.0
[37] hms_1.1.1 digest_0.6.28 stringi_1.7.5 grid_4.0.3
[41] cli_3.1.0 tools_4.0.3 magrittr_2.0.1 sass_0.4.0
[45] crayon_1.4.2 pkgconfig_2.0.3 ellipsis_0.3.2 xml2_1.3.2
[49] reprex_2.0.1 lubridate_1.8.0 rstudioapi_0.13 assertthat_0.2.1
[53] rmarkdown_2.11 httr_1.4.2 R6_2.5.1 compiler_4.0.3
Preliminary work¶
Documentation on visNetwork can be found here.
Data importation¶
All the files imported here can be found on Genotoul, in /work2/project/regenet/results/multi/abc.model/Nasser2021.
The full code itself is available here: preliminary_analysis_v10.Rmd.
rm(list = ls())
# wd = 'data/' wd =
# '/home/hoellinger/Documents/INSERM/shared/automne_2021/networks_hemochromatosis/data/'
wd = "/home/thoellinger/Documents/shared/automne_2021/networks_hemochromatosis/data/"
egfile = paste(wd, "Nasser2021ABCPredictions.liver_and_intestine.all_putative_enhancers.merged_enhancers.sorted.uniques_eg.bedpe",
sep = "")
unmerged_efile = paste(wd, "list_all_enhancers.bed", sep = "")
merged_efile = paste(wd, "list_all_enhancers.merged.bed", sep = "")
############# Enhancers # The only reasons all enhancers are loaded is to
############# compute a few summary statistics.
e = as.data.frame(read.table(unmerged_efile, sep = "\t"))
me = as.data.frame(read.table(merged_efile, sep = "\t"))
############ E-G list #
eg = as.data.frame(read.table(egfile, header = T, sep = "\t"))
############### known genes #
gene_list = c("HFE", "TFR2", "HFE2", "HAMP", "SLC40A1", "BMP6", "TMPRSS6", "TFRC",
"SLC11A2", "CYBRD1", "NEO1", "CIAPIN1", "SLC39A14")
# genes known to be causally involved in hemochromatosis: 'HFE', 'TFR2',
# 'HFE2', 'HAMP', 'SLC40A1', 'BMP6' (the other genes are involved in iron
# metabolism regulation) genes expressed in the liver: 'HFE', 'TFR2', 'HFE2',
# 'HAMP', 'SLC40A1', 'BMP6', 'TMPRSS6', 'TFRC' genes expressed in intestine:
# 'SLC40A1', 'SLC11A2', 'CYBRD1', 'NEO1', 'CIAPIN1'
Data preprocessing¶
New columns¶
We will need two more columns containing the following:
eg$biosamples.uniq = unlist(lapply(eg$biosamples, function(x) paste(unlist(sort(unique(strsplit(x,
",")[[1]]))), collapse = ",")))
eg$tissues.uniq = unlist(lapply(eg$tissues, function(x) paste(unlist(sort(unique(strsplit(x,
",")[[1]]))), collapse = ",")))
Conversion to factors¶
This has to be done AFTER the creation of new columns done above:
to_factor_cols = c("chrom1", "chrom2", "name", "strand1", "strand2", "gene", "biosamples",
"tissues", "biosamples.uniq", "tissues.uniq")
eg[to_factor_cols] = lapply(eg[to_factor_cols], factor)
Exploration¶
Summary statistics on enhancer lists¶
length(e[, 1]) # list of all enhancers
[1] 2463310
length(me[, 1]) # list of all merged enhancers (such that none of those merged enhancers are overlapping)
[1] 269254
Warning: we shall pay attention to the fact that among those 269,254 merged putative enhancers, 85,937 (32%) do not overlap any ccRE-ELS, and only 112,356 enhancers (42%) overlap exactly one ccRE-ELS (but the latter behavior is expected because of the merging process). Contrariwise, only 25,709 of those enhancers (9%) do not overlap any ccRE (ie candidate regulatory element not necessarily with Enhancer-Like-Signature), suggesting that many of those 269,254 merged putative enhancers might not be real enhancers (ie with both high DNase and high H3K27ac signal in one or more of the ENCODE biosamples used to defined ccRE), but CTCT-only, promoters or DNase-H3K4me2 regions.
Nevertheless, we chose to use the list of 269,254 putative enhancers for consistency in our analysis when it comes to compare results when removing or adding a new biosamples / tissues (so that the list of enhancers does not change in the process). In the future, we might take some time to compare what we would have obtained otherwise.
e$length = abs(e$V3 - e$V2)
me$length = abs(me$V3 - me$V2)
summary(e$length)
Min. 1st Qu. Median Mean 3rd Qu. Max.
200.0 200.0 308.0 481.7 637.0 6991.0
summary(me$length)
Min. 1st Qu. Median Mean 3rd Qu. Max.
200.0 288.0 631.0 807.8 1078.0 11616.0
E-G pairs¶
We extract the subsample of the E-G bedpe input list, where genes are contained in our list of genes involved either directly in hemochromatosis or in iron metabolism. In the variable name, "dist0" stands for "distance is 0 between the genes in eg_dist0 and the list of initial genes".
eg_dist0 = eg[eg$gene %in% gene_list, ]
length(eg_dist0$name)
[1] 137
length(unique(eg_dist0$gene))
[1] 13
table(eg_dist0$tissues.uniq)
intestine intestine,liver liver
36 21 80
Genes¶
Chromosomes where the genes are located:
HFE TFR2 HFE2 HAMP SLC40A1 BMP6 TMPRSS6 TFRC
"chr6" "chr7" "chr1" "chr19" "chr2" "chr6" "chr22" "chr3"
SLC11A2 CYBRD1 NEO1 CIAPIN1 SLC39A14
"chr12" "chr2" "chr15" "chr16" "chr8"
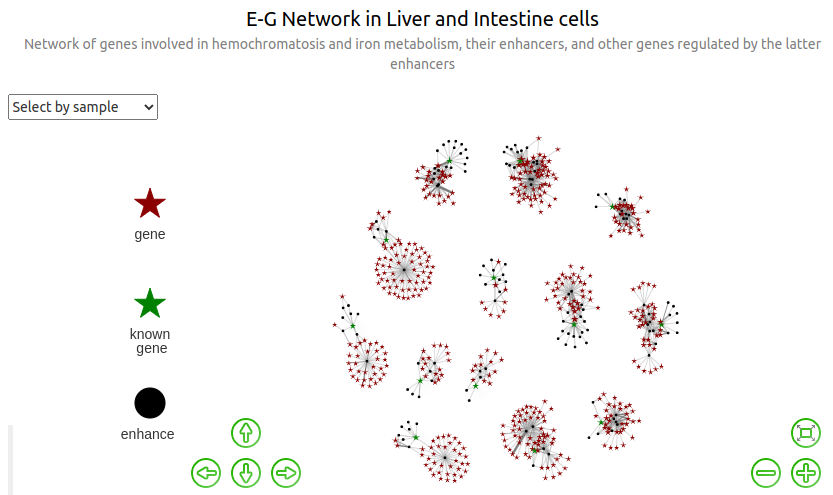
Networks¶
Note: in all subsequent graphs, size of nodes of type "gene" (and not "known_gene", for which the size is fixed) is proportional to the number of distinct enhancers regulating them.
Find all genes regulated by the initial enhancers¶
The "initial enhancers" are the enhancers involved in eg_dist0, ie all the enhancers regulating the initial genes in gene_list.
genes is the same as gene_list (strictly speaking, genes is the subset of gene_list for which we have data), and enhancers is the list of enhancers regulating those initial known genes.
enhancers = unique(paste(eg_dist0$chrom1, ":", eg_dist0$start1, "-", eg_dist0$end1,
sep = ""))
genes = unique(eg_dist0$gene)
Now we compute the list of all genes regulated by enhancers in eg_dist0. Specifically, we extract, from the full E-G list eg, the list eg_dist1 containing only the enhancers-genes pairs for which the gene G is regulated by any of the enhancers regulating a gene in gene_list ("dist1" stands for "distance is at most 1 between the genes in eg_dist1 and the list of initial genes").
eg_enhancers_id = data.frame(source = paste(eg$chrom1, ":", eg$start1, "-", eg$end1,
sep = ""), eg[, 4:22]) # same as eg but columns 1-3 have been concatenated to make unique enhancers id
eg_dist1 = eg_enhancers_id[eg_enhancers_id$source %in% paste(eg_dist0$chrom1, ":",
eg_dist0$start1, "-", eg_dist0$end1, sep = ""), ]
eg_dist1$from = lapply(eg_dist1$source, function(x) unique(as.character(eg_dist0[paste(eg_dist0$chrom1,
":", eg_dist0$start1, "-", eg_dist0$end1, sep = "") == x, "gene"])))
eg_dist1$ABC.IE = left_join(data.frame(name = paste(eg_dist1$source, eg_dist1$from,
sep = "::")), eg_enhancers_id, by = "name")$ABC.max # max ABC score of the I-E pair (initialGene-Enhancer) corresponding to the E-G pair
genes_dist1 = unique(eg_dist1$gene)
genes_dist1 is the list of genes regulated by enhancers.
Compute "ABC product", ie the product of the ABC scores of: - the initial gene - enhancer pair (I-E) - the enhancer - gene pair (E-G)
eg_dist1$ABC.product = eg_dist1$ABC.max * eg_dist1$ABC.IE
eg_dist1$ABC.product = eg_dist1$ABC.product/max(eg_dist1$ABC.product)
print(min(eg_dist1$ABC.product))
[1] 0.003458878
print(median(eg_dist1$ABC.product))
[1] 0.01070413
print(mean(eg_dist1$ABC.product))
[1] 0.02172209
print(quantile(eg_dist1$ABC.product, c(0.1, 0.6, 0.8, 0.9)))
10% 60% 80% 90%
0.004902703 0.013383311 0.027607616 0.044685971
print(max(eg_dist1$ABC.product))
[1] 1
eg_dist1$ABC.product.label = 1
eg_dist1[eg_dist1$ABC.product >= median(eg_dist1$ABC.product), ]$ABC.product.label = 2
eg_dist1[eg_dist1$ABC.product >= quantile(eg_dist1$ABC.product, 0.9)[[1]], ]$ABC.product.label = 3
table(eg_dist1$ABC.product.label)
1 2 3
673 539 135
In the following cell we create genes_dist1.more which is in a well-suited format for further "concatenation" with inferences made with other type of data (CHiC or QTL -based).
genes_dist1.more = eg_dist1 %>%
group_by(gene) %>%
mutate(ABC.sources = paste0(source, collapse = ",")) %>%
mutate(ABC.count = length(str_split(ABC.sources, ",")[[1]])) %>%
group_by(gene) %>%
slice(which.max(ABC.product.label)) %>%
# slice_max(ABC.product.label) %>%
select(gene, ABC.sources, ABC.product.label, ABC.count, from)
genes_dist1.more = subset(genes_dist1.more, !(genes_dist1.more$gene %in% genes))
genes_dist1.more$from = as.character(genes_dist1.more$from)
genes_dist1.more
# A tibble: 444 × 5
# Groups: gene [444]
gene ABC.sources ABC.product.lab… ABC.count from
<fct> <chr> <dbl> <int> <chr>
1 AAAS chr12:53318068-53321694 1 1 SLC11…
2 ABT1 chr6:24719303-24722493 1 1 HFE
3 ACOT13 chr6:24719303-24722493,chr6:25991… 3 2 HFE
4 ACTL6B chr7:100166895-100168233,chr7:100… 2 2 TFR2
5 ACVR1B chr12:53318068-53321694 2 1 SLC11…
6 ACVRL1 chr12:53318068-53321694 2 1 SLC11…
7 ADAM28 chr8:22417293-22420339 2 1 SLC39…
8 ADAMDEC1 chr8:22417293-22420339 2 1 SLC39…
9 ADGRG1 chr16:56639960-56645831,chr16:573… 3 7 CIAPI…
10 ADGRG3 chr16:56639960-56645831,chr16:573… 2 4 CIAPI…
# … with 434 more rows
We re-arrange eg_dist1 as an edge list edges_list_dist1, which will be more suitable to later construct the edges list for visualization as a graph.
edges_list_dist1 = data.frame(source = eg_dist1$source, target = eg_dist1$gene, ABC.mean.x100 = floor(100 *
eg_dist1$ABC.mean), ABC.max.x100 = floor(100 * eg_dist1$ABC.max), tissues = eg_dist1$tissues.uniq,
distance_kb = floor(eg_dist1$original_distance.mean/1000), inv_dist = 1/(eg_dist1$original_distance.mean +
1), rescaled_log_inv_dist = 1 - min(log(1/(eg_dist1$original_distance.mean +
1))) + log(1/(eg_dist1$original_distance.mean + 1)))
edges_list_dist1
Below we compute the list of (colored) nodes required to compute our graphs, a node corresponding either to an enhancer or a gene. To that purpose, we need first to construct 2 tables:
- gname.tissue contains, for each unique gene involved in eg_dist1, the comma-separated list of tissues in which it appears in eg_dist1
- enhancer.tissue contains, for each unique enhancer involved in eg_dist1, the comma-separated list of tissues in which it appears in eg_dist1
For more details on the Reduce function and its application to our case, see for instance: https://stackoverflow.com/questions/60592775/how-to-apply-reduce-to-groups-based-on-columns-of-a-data-frame
gname.tissue = unique(eg_dist1[c("gene", "tissues.uniq")])
gname.tissue$tissues.uniq = as.character(gname.tissue$tissues.uniq)
gname.tissue = gname.tissue %>%
group_by(gene) %>%
mutate(tissues = Reduce(function(x, y) {
paste(unlist(sort(unique(c(strsplit(x, ",")[[1]], strsplit(y, ",")[[1]])))),
collapse = ",")
}, tissues.uniq))
gname.tissue = unique(gname.tissue[c(1, 3)])
enhancer.tissue = unique(eg_dist1[c("source", "tissues.uniq")])
enhancer.tissue$tissues.uniq = as.character(enhancer.tissue$tissues.uniq)
enhancer.tissue = enhancer.tissue %>%
group_by(source) %>%
mutate(tissues = Reduce(function(x, y) {
paste(unlist(sort(unique(c(strsplit(x, ",")[[1]], strsplit(y, ",")[[1]])))),
collapse = ",")
}, tissues.uniq))
enhancer.tissue = unique(enhancer.tissue[c(1, 3)])
gname.tissue
# A tibble: 457 × 2
# Groups: gene [457]
gene tissues
<fct> <chr>
1 PDE4DIP intestine,liver
2 LOC100996724 intestine,liver
3 SEC22B intestine,liver
4 NOTCH2NL intestine,liver
5 NBPF10 intestine,liver
6 LINC01719 intestine,liver
7 HFE2 liver
8 TXNIP intestine,liver
9 POLR3GL intestine,liver
10 ANKRD34A intestine,liver
# … with 447 more rows
enhancer.tissue
# A tibble: 137 × 2
# Groups: source [137]
source tissues
<chr> <chr>
1 chr1:144930398-144933456 intestine,liver
2 chr1:145394876-145400153 intestine,liver
3 chr1:145413923-145415673 liver
4 chr1:145421224-145422239 liver
5 chr1:145426950-145428636 intestine,liver
6 chr1:145434950-145435719 intestine,liver
7 chr1:145442141-145445254 intestine,liver
8 chr1:145451402-145451862 liver
9 chr1:145454003-145457954 intestine,liver
10 chr1:145473958-145474912 intestine,liver
# … with 127 more rows
So now we can compute the list nodes_dist1 of (colored) nodes required for our graphs. There are 3 types of nodes: enhancer, known_gene and (unknown) gene.
nodes_dist1 = full_join(data.frame(label = enhancer.tissue$source, sample = enhancer.tissue$tissues,
group = "enhancer"), data.frame(label = gname.tissue$gene, sample = gname.tissue$tissues,
group = "gene")) %>%
rowid_to_column("id")
nodes_dist1[nodes_dist1$label %in% genes, ]$group = "known_gene"
table(nodes_dist1$sample)
intestine intestine,liver liver
135 290 169
table(nodes_dist1$group)
enhancer gene known_gene
137 444 13
In the list of edges of the graph, edges_dist1, the sample column indicates in which family of tissues (liver, intestine or both) each E-G pair has been found.
table(edges_dist1$sample)
intestine intestine,liver liver
420 321 606
We see that 1221 unique E-G pairs found, 46% are specific to liver, 29% are specific to intestine and 24% are shared between intestine and liver.
We add to nodes_dist1 a column d_in for plotting purpose: it contains 1 for each node of type enhancer, the number of incoming enhancers for each node of type gene, and the max of the latter for each node of type known_gene.
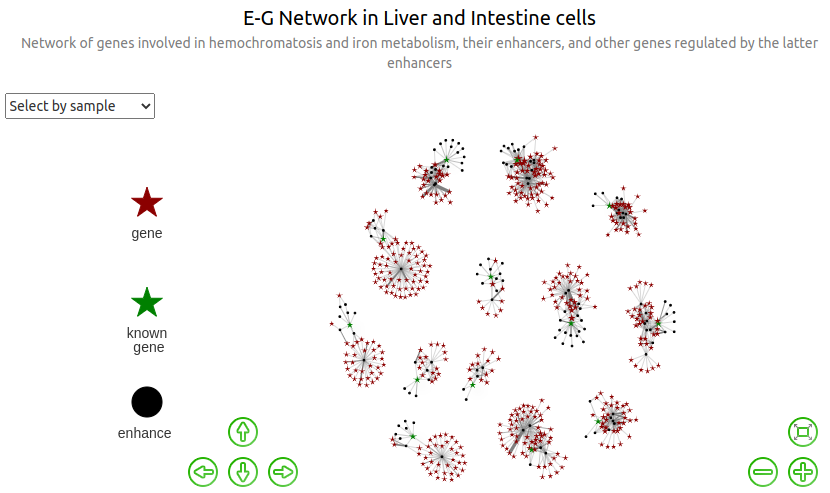
Edge weight based on ABC score¶
ABC.mean¶
In this graph, the weights of the edges are proportional to the corresponding mean ABC scores (averaged over all the instances of the E-G pairs found in the different biosamples. Not that, for a given biosample, the "ABC.score" column contains ABC scores that have already been averaged once when merging the enhancers).
We see that none of the genes connected through enhancers to different genes among the initial list of 13 genes ; are connected to more than 1 of them ; ie we still have 13 connected compounds. Yet, most of the initial genes are on different chromosomes, so this is a completely expected behavior.
Also note that one may observe that when selecting a sample, all edges connected to enhancers included in the selected groups are colored (ie are in darker grey than the others), and not all edges connected to genes included in the selected group. The actual reason is that the graph is considered as an oriented graph, resulting in a distinction between entering and outgoing edges. Namely, for each selected nodes, all outgoing edges are colored ; whereas regarding entering edges, only those coming from another selected nodes are colored. In our case, all edges connected to any enhancer are considered as outgoing edges, and all edges connected to any gene are considered as entering edges ; which explains what we observe. Unfortunately there is nothing we can do with the
visNetworkpackage to change this behavior.
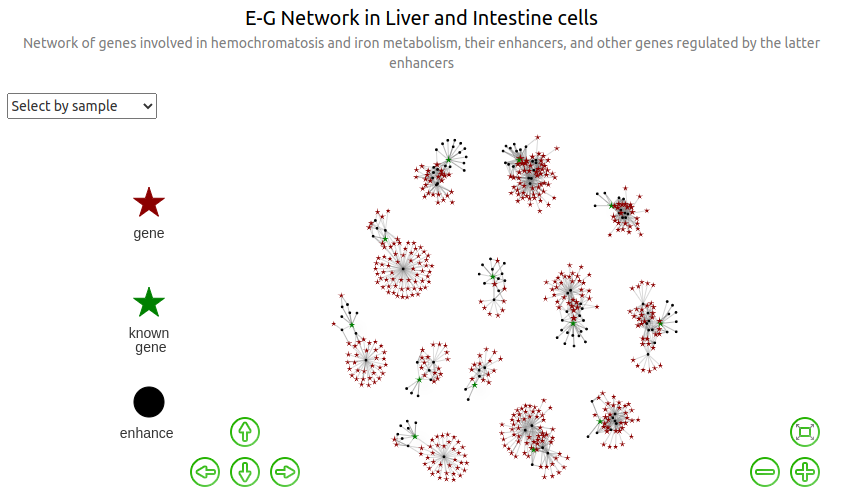
ABC.max¶
In this graph, the weights of the edges are proportional to the corresponding max ABC scores (over all the instances of the E-G pairs found in the different biosamples. Not that, for a given biosample, the "ABC.score" column contains ABC scores that have already been averaged once when merging the enhancers).
Edge weight based on distance¶
Width proportional to mean distanc. For each enhancer-gene pair $E$-$G$, the distance indicated in the eg dataframe as original_distance.mean is the average distance $E_b$-$G$ over the biosamples $b$ (in base pairs), where $E_b$ is the original enhancer in biosample $b$ which has been replaced by its overlapping merged enhancer $E$. Here, distance_kb is the very same quantity but expressed in kb.
Width proportional to inverse distance:
Width proportional to rescaled and translated log inverse distance:
Results¶
The list of the initial genes + all genes regulated by enhancers regulating the 13 initial genes, can be found here: /work2/project/regenet/workspace/thoellinger/shared/2022/networks_hemochromatosis/results/new_genes_v7.list.
write.table(genes_dist1, file = "results/new_genes_v7.list", quote = FALSE, sep = "\t",
row.names = F, col.names = F)
The list all genes regulated by enhancers regulating the 13 initial genes, with useful info, can be found here: /work2/project/regenet/workspace/thoellinger/shared/2022/networks_hemochromatosis/results/new_genes_abc_v9_more_info.list.
write.table(genes_dist1.more, file = "results/new_genes_abc_v9_more_info.list", quote = FALSE,
sep = "\t", row.names = F, col.names = T)
For each one of the 13 initial genes, the list of genes regulated by enhancers regulating that initial gene, can be found here:
for (gene in genes) {
current_gene_dist1 = unique(eg_dist1[eg_dist1$from == gene, "gene"])
write.table(current_gene_dist1, file = paste("results/separate/", gene, ".list",
sep = ""), quote = FALSE, sep = "\t", row.names = F, col.names = F)
}
The list of the enhancers regulating the 13 initial genes, can be found here: /work2/project/regenet/workspace/thoellinger/shared/2022/networks_hemochromatosis/results/enhancers.list
write.table(enhancers, file = "results/enhancers.list", quote = FALSE, sep = "\t",
row.names = F, col.names = F)
The list of E-G pairs involving the 13 initial genes, can be found here: /work2/project/regenet/workspace/thoellinger/shared/2022/networks_hemochromatosis/results/eg_dist0.bedpe
write.table(eg_dist0, file = "results/eg_dist0.bedpe", quote = FALSE, sep = "\t",
row.names = F, col.names = F)
The list of the enhancers regulating the 457 initial+new genes, can be found here: /work2/project/regenet/workspace/thoellinger/shared/2022/networks_hemochromatosis/results/enhancers.new_genes.list
eg_dist2 = eg[eg$gene %in% genes_dist1, ]
enhancers.new_genes = unique(paste(eg_dist2$chrom1, ":", eg_dist2$start1, "-", eg_dist2$end1,
sep = ""))
write.table(enhancers.new_genes, file = "results/enhancers.new_genes.list", quote = FALSE,
sep = "\t", row.names = F, col.names = F)
The list of E-G pairs involving the 457 initial+new genes, can be found here: /work2/project/regenet/workspace/thoellinger/shared/2022/networks_hemochromatosis/results/eg_dist2.bedpe
write.table(eg_dist2, file = "results/eg_dist2.bedpe", quote = FALSE, sep = "\t",
row.names = F, col.names = F)
Appendix¶
Tables¶
Table of the initial ABC-predicted E-G pairs involving the initial genes¶
options(max.print = 1000)
eg_dist0
chrom1 start1 end1 chrom2 start2 end2
7686 chr1 144930398 144933456 chr1 145413190 145413190
7781 chr1 145394876 145400153 chr1 145413190 145413190
7797 chr1 145413923 145415673 chr1 145413190 145413190
7798 chr1 145421224 145422239 chr1 145413190 145413190
7799 chr1 145426950 145428636 chr1 145413190 145413190
7801 chr1 145434950 145435719 chr1 145413190 145413190
7815 chr1 145442141 145445254 chr1 145413190 145413190
7831 chr1 145451402 145451862 chr1 145413190 145413190
7846 chr1 145454003 145457954 chr1 145413190 145413190
7867 chr1 145473958 145474912 chr1 145413190 145413190
7894 chr1 145541765 145543880 chr1 145413190 145413190
30929 chr12 51158462 51159633 chr12 51420199 51420199
30935 chr12 51326262 51329574 chr12 51420199 51420199
30939 chr12 51417885 51419543 chr12 51420199 51420199
30946 chr12 51421716 51423284 chr12 51420199 51420199
31061 chr12 53318068 53321694 chr12 51420199 51420199
46386 chr15 73074997 73077510 chr15 73344824 73344824
46391 chr15 73320830 73321661 chr15 73344824 73344824
46392 chr15 73326079 73327042 chr15 73344824 73344824
46393 chr15 73345478 73345779 chr15 73344824 73344824
46394 chr15 73346002 73346202 chr15 73344824 73344824
46395 chr15 73346837 73347246 chr15 73344824 73344824
46398 chr15 73401919 73403010 chr15 73344824 73344824
46488 chr15 74906565 74909246 chr15 73344824 73344824
51946 chr16 56639960 56645831 chr16 57481440 57481440
52046 chr16 57298869 57301500 chr16 57481440 57481440
52075 chr16 57302558 57303592 chr16 57481440 57481440
52123 chr16 57333210 57335328 chr16 57481440 57481440
52149 chr16 57337937 57338465 chr16 57481440 57481440
52156 chr16 57417430 57418771 chr16 57481440 57481440
52160 chr16 57451676 57452165 chr16 57481440 57481440
52164 chr16 57452493 57453437 chr16 57481440 57481440
52166 chr16 57466172 57466712 chr16 57481440 57481440
52168 chr16 57477983 57478427 chr16 57481440 57481440
52171 chr16 57504477 57506928 chr16 57481440 57481440
52177 chr16 57512427 57514785 chr16 57481440 57481440
52193 chr16 57553700 57554263 chr16 57481440 57481440
52215 chr16 57653300 57655255 chr16 57481440 57481440
52238 chr16 57669027 57670023 chr16 57481440 57481440
52283 chr16 57830970 57833098 chr16 57481440 57481440
52345 chr16 57925806 57928939 chr16 57481440 57481440
68172 chr19 35757791 35760841 chr19 35773248 35773248
68173 chr19 35768288 35769137 chr19 35773248 35773248
68174 chr19 35769477 35770158 chr19 35773248 35773248
68175 chr19 35770384 35772541 chr19 35773248 35773248
name strand1 strand2 gene
7686 chr1:144930398-144933456::HFE2 . . HFE2
7781 chr1:145394876-145400153::HFE2 . . HFE2
7797 chr1:145413923-145415673::HFE2 . . HFE2
7798 chr1:145421224-145422239::HFE2 . . HFE2
7799 chr1:145426950-145428636::HFE2 . . HFE2
7801 chr1:145434950-145435719::HFE2 . . HFE2
7815 chr1:145442141-145445254::HFE2 . . HFE2
7831 chr1:145451402-145451862::HFE2 . . HFE2
7846 chr1:145454003-145457954::HFE2 . . HFE2
7867 chr1:145473958-145474912::HFE2 . . HFE2
7894 chr1:145541765-145543880::HFE2 . . HFE2
30929 chr12:51158462-51159633::SLC11A2 . . SLC11A2
30935 chr12:51326262-51329574::SLC11A2 . . SLC11A2
30939 chr12:51417885-51419543::SLC11A2 . . SLC11A2
30946 chr12:51421716-51423284::SLC11A2 . . SLC11A2
31061 chr12:53318068-53321694::SLC11A2 . . SLC11A2
46386 chr15:73074997-73077510::NEO1 . . NEO1
46391 chr15:73320830-73321661::NEO1 . . NEO1
46392 chr15:73326079-73327042::NEO1 . . NEO1
46393 chr15:73345478-73345779::NEO1 . . NEO1
46394 chr15:73346002-73346202::NEO1 . . NEO1
46395 chr15:73346837-73347246::NEO1 . . NEO1
46398 chr15:73401919-73403010::NEO1 . . NEO1
46488 chr15:74906565-74909246::NEO1 . . NEO1
51946 chr16:56639960-56645831::CIAPIN1 . . CIAPIN1
52046 chr16:57298869-57301500::CIAPIN1 . . CIAPIN1
52075 chr16:57302558-57303592::CIAPIN1 . . CIAPIN1
52123 chr16:57333210-57335328::CIAPIN1 . . CIAPIN1
52149 chr16:57337937-57338465::CIAPIN1 . . CIAPIN1
52156 chr16:57417430-57418771::CIAPIN1 . . CIAPIN1
52160 chr16:57451676-57452165::CIAPIN1 . . CIAPIN1
52164 chr16:57452493-57453437::CIAPIN1 . . CIAPIN1
52166 chr16:57466172-57466712::CIAPIN1 . . CIAPIN1
52168 chr16:57477983-57478427::CIAPIN1 . . CIAPIN1
52171 chr16:57504477-57506928::CIAPIN1 . . CIAPIN1
52177 chr16:57512427-57514785::CIAPIN1 . . CIAPIN1
52193 chr16:57553700-57554263::CIAPIN1 . . CIAPIN1
52215 chr16:57653300-57655255::CIAPIN1 . . CIAPIN1
52238 chr16:57669027-57670023::CIAPIN1 . . CIAPIN1
52283 chr16:57830970-57833098::CIAPIN1 . . CIAPIN1
52345 chr16:57925806-57928939::CIAPIN1 . . CIAPIN1
68172 chr19:35757791-35760841::HAMP . . HAMP
68173 chr19:35768288-35769137::HAMP . . HAMP
68174 chr19:35769477-35770158::HAMP . . HAMP
68175 chr19:35770384-35772541::HAMP . . HAMP
ABC.scores
7686 0.028761000000000002,0.02026
7781 0.044185
7797 0.062645
7798 0.015668
7799 0.02574
7801 0.028239999999999998,0.015686000000000002
7815 0.062419
7831 0.021353999999999998
7846 0.028807999999999997,0.0489073
7867 0.039256,0.021347
7894 0.045182,0.01541
30929 0.018211
30935 0.015196000000000001,0.022472
30939 0.05407000000000001,0.055278999999999995,0.065178,0.062988
30946 0.138303,0.145403
31061 0.018961000000000002
46386 0.024743
46391 0.05282100000000001
46392 0.042369
46393 0.022713999999999998
46394 0.015252000000000002
46395 0.02547,0.029026
46398 0.020356,0.027807
46488 0.01845
51946 0.018703
52046 0.0205155
52075 0.030206999999999998,0.03244
52123 0.052578,0.088226,0.024952000000000002,0.06717999999999999
52149 0.018795
52156 0.0161
52160 0.020324000000000002
52164 0.015609999999999999
52166 0.016451,0.015881
52168 0.034379
52171 0.016694,0.0385055,0.021072999999999998,0.022296,0.024221
52177 0.025434000000000002,0.031333,0.024978999999999998,0.022293
52193 0.016816
52215 0.017281,0.024939
52238 0.017934000000000002,0.017163
52283 0.017979
52345 0.017008000000000002,0.015305000000000001
68172 0.020316999999999998
68173 0.017357
68174 0.015806999999999998
68175 0.06381,0.023445,0.065302
biosamples
7686 liver-ENCODE,hepatocyte-ENCODE
7781 liver-ENCODE
7797 hepatocyte-ENCODE
7798 hepatocyte-ENCODE
7799 liver-ENCODE
7801 liver-ENCODE,hepatocyte-ENCODE
7815 liver-ENCODE
7831 liver-ENCODE
7846 hepatocyte-ENCODE,liver-ENCODE
7867 hepatocyte-ENCODE,liver-ENCODE
7894 hepatocyte-ENCODE,liver-ENCODE
30929 hepatocyte-ENCODE
30935 liver-ENCODE,HepG2-Roadmap
30939 small_intestine_fetal-Roadmap,HepG2-Roadmap,hepatocyte-ENCODE,liver-ENCODE
30946 small_intestine_fetal-Roadmap,large_intestine_fetal-Roadmap
31061 large_intestine_fetal-Roadmap
46386 hepatocyte-ENCODE
46391 large_intestine_fetal-Roadmap
46392 large_intestine_fetal-Roadmap
46393 HepG2-Roadmap
46394 small_intestine_fetal-Roadmap
46395 large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
46398 small_intestine_fetal-Roadmap,large_intestine_fetal-Roadmap
46488 hepatocyte-ENCODE
51946 liver-ENCODE
52046 small_intestine_fetal-Roadmap
52075 large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
52123 liver-ENCODE,large_intestine_fetal-Roadmap,HepG2-Roadmap,small_intestine_fetal-Roadmap
52149 liver-ENCODE
52156 large_intestine_fetal-Roadmap
52160 liver-ENCODE
52164 liver-ENCODE
52166 large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
52168 hepatocyte-ENCODE
52171 small_intestine_fetal-Roadmap,hepatocyte-ENCODE,large_intestine_fetal-Roadmap,liver-ENCODE,HepG2-Roadmap
52177 small_intestine_fetal-Roadmap,hepatocyte-ENCODE,HepG2-Roadmap,large_intestine_fetal-Roadmap
52193 hepatocyte-ENCODE
52215 small_intestine_fetal-Roadmap,large_intestine_fetal-Roadmap
52238 large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
52283 liver-ENCODE
52345 HepG2-Roadmap,liver-ENCODE
68172 liver-ENCODE
68173 hepatocyte-ENCODE
68174 HepG2-Roadmap
68175 HepG2-Roadmap,liver-ENCODE,hepatocyte-ENCODE
original_distances
7686 481207.0,480853.0
7781 15386.0
7797 1252.0
7798 8577.0
7799 14427.0
7801 22160.0,22318.0
7815 30687.0
7831 38465.5
7846 44235.5,44088.0
7867 61135.0,61186.0
7894 129732.5,129661.0
30929 261301.5
30935 91637.0,92887.0
30939 1340.5,1461.0,1895.5,1741.0
30946 2216.0,2198.0
31061 1900020.0
46386 267544.0
46391 23578.5
46392 18400.5
46393 855.0
46394 1278.0
46395 2205.0,2198.0
46398 57556.0,57790.0
46488 1563131.0
51946 840504.5
52046 181096.0
52075 178475.5,178416.5
52123 147262.0,147349.0,147375.0,147212.0
52149 143184.5
52156 63738.0
52160 29562.0
52164 28344.5
52166 14998.0,14968.0
52168 3235.0
52171 24545.0,24996.5,24548.0,23673.5,24795.5
52177 32869.0,32909.0,32686.5,32869.0
52193 72489.5
52215 172594.5,172494.0
52238 188158.5,188051.0
52283 350469.5
52345 446763.0,446265.0
68172 14913.0
68173 4328.0
68174 3303.0
68175 2157.0,1050.0,1856.5
tissues ABC.mean ABC.min ABC.max
7686 liver,liver 0.02451050 0.0202600 0.0287610
7781 liver 0.04418500 0.0441850 0.0441850
7797 liver 0.06264500 0.0626450 0.0626450
7798 liver 0.01566800 0.0156680 0.0156680
7799 liver 0.02574000 0.0257400 0.0257400
7801 liver,liver 0.02196300 0.0156860 0.0282400
7815 liver 0.06241900 0.0624190 0.0624190
7831 liver 0.02135400 0.0213540 0.0213540
7846 liver,liver 0.03885765 0.0288080 0.0489073
7867 liver,liver 0.03030150 0.0213470 0.0392560
7894 liver,liver 0.03029600 0.0154100 0.0451820
30929 liver 0.01821100 0.0182110 0.0182110
30935 liver,liver 0.01883400 0.0151960 0.0224720
30939 intestine,liver,liver,liver 0.05937875 0.0540700 0.0651780
30946 intestine,intestine 0.14185300 0.1383030 0.1454030
31061 intestine 0.01896100 0.0189610 0.0189610
46386 liver 0.02474300 0.0247430 0.0247430
46391 intestine 0.05282100 0.0528210 0.0528210
46392 intestine 0.04236900 0.0423690 0.0423690
46393 liver 0.02271400 0.0227140 0.0227140
46394 intestine 0.01525200 0.0152520 0.0152520
46395 intestine,intestine 0.02724800 0.0254700 0.0290260
46398 intestine,intestine 0.02408150 0.0203560 0.0278070
46488 liver 0.01845000 0.0184500 0.0184500
51946 liver 0.01870300 0.0187030 0.0187030
52046 intestine 0.02051550 0.0205155 0.0205155
52075 intestine,intestine 0.03132350 0.0302070 0.0324400
52123 liver,intestine,liver,intestine 0.05823400 0.0249520 0.0882260
52149 liver 0.01879500 0.0187950 0.0187950
52156 intestine 0.01610000 0.0161000 0.0161000
52160 liver 0.02032400 0.0203240 0.0203240
52164 liver 0.01561000 0.0156100 0.0156100
52166 intestine,intestine 0.01616600 0.0158810 0.0164510
52168 liver 0.03437900 0.0343790 0.0343790
52171 intestine,liver,intestine,liver,liver 0.02455790 0.0166940 0.0385055
52177 intestine,liver,liver,intestine 0.02600975 0.0222930 0.0313330
52193 liver 0.01681600 0.0168160 0.0168160
52215 intestine,intestine 0.02111000 0.0172810 0.0249390
52238 intestine,intestine 0.01754850 0.0171630 0.0179340
52283 liver 0.01797900 0.0179790 0.0179790
52345 liver,liver 0.01615650 0.0153050 0.0170080
68172 liver 0.02031700 0.0203170 0.0203170
68173 liver 0.01735700 0.0173570 0.0173570
68174 liver 0.01580700 0.0158070 0.0158070
68175 liver,liver,liver 0.05085233 0.0234450 0.0653020
original_distance.mean original_distance.min original_distance.max
7686 481030.000 480853.0 481207.0
7781 15386.000 15386.0 15386.0
7797 1252.000 1252.0 1252.0
7798 8577.000 8577.0 8577.0
7799 14427.000 14427.0 14427.0
7801 22239.000 22160.0 22318.0
7815 30687.000 30687.0 30687.0
7831 38465.500 38465.5 38465.5
7846 44161.750 44088.0 44235.5
7867 61160.500 61135.0 61186.0
7894 129696.750 129661.0 129732.5
30929 261301.500 261301.5 261301.5
30935 92262.000 91637.0 92887.0
30939 1609.500 1340.5 1895.5
30946 2207.000 2198.0 2216.0
31061 1900020.000 1900020.0 1900020.0
46386 267544.000 267544.0 267544.0
46391 23578.500 23578.5 23578.5
46392 18400.500 18400.5 18400.5
46393 855.000 855.0 855.0
46394 1278.000 1278.0 1278.0
46395 2201.500 2198.0 2205.0
46398 57673.000 57556.0 57790.0
46488 1563131.000 1563131.0 1563131.0
51946 840504.500 840504.5 840504.5
52046 181096.000 181096.0 181096.0
52075 178446.000 178416.5 178475.5
52123 147299.500 147212.0 147375.0
52149 143184.500 143184.5 143184.5
52156 63738.000 63738.0 63738.0
52160 29562.000 29562.0 29562.0
52164 28344.500 28344.5 28344.5
52166 14983.000 14968.0 14998.0
52168 3235.000 3235.0 3235.0
52171 24511.700 23673.5 24996.5
52177 32833.375 32686.5 32909.0
52193 72489.500 72489.5 72489.5
52215 172544.250 172494.0 172594.5
52238 188104.750 188051.0 188158.5
52283 350469.500 350469.5 350469.5
52345 446514.000 446265.0 446763.0
68172 14913.000 14913.0 14913.0
68173 4328.000 4328.0 4328.0
68174 3303.000 3303.0 3303.0
68175 1687.833 1050.0 2157.0
biosamples.uniq
7686 hepatocyte-ENCODE,liver-ENCODE
7781 liver-ENCODE
7797 hepatocyte-ENCODE
7798 hepatocyte-ENCODE
7799 liver-ENCODE
7801 hepatocyte-ENCODE,liver-ENCODE
7815 liver-ENCODE
7831 liver-ENCODE
7846 hepatocyte-ENCODE,liver-ENCODE
7867 hepatocyte-ENCODE,liver-ENCODE
7894 hepatocyte-ENCODE,liver-ENCODE
30929 hepatocyte-ENCODE
30935 HepG2-Roadmap,liver-ENCODE
30939 hepatocyte-ENCODE,HepG2-Roadmap,liver-ENCODE,small_intestine_fetal-Roadmap
30946 large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
31061 large_intestine_fetal-Roadmap
46386 hepatocyte-ENCODE
46391 large_intestine_fetal-Roadmap
46392 large_intestine_fetal-Roadmap
46393 HepG2-Roadmap
46394 small_intestine_fetal-Roadmap
46395 large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
46398 large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
46488 hepatocyte-ENCODE
51946 liver-ENCODE
52046 small_intestine_fetal-Roadmap
52075 large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
52123 HepG2-Roadmap,large_intestine_fetal-Roadmap,liver-ENCODE,small_intestine_fetal-Roadmap
52149 liver-ENCODE
52156 large_intestine_fetal-Roadmap
52160 liver-ENCODE
52164 liver-ENCODE
52166 large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
52168 hepatocyte-ENCODE
52171 hepatocyte-ENCODE,HepG2-Roadmap,large_intestine_fetal-Roadmap,liver-ENCODE,small_intestine_fetal-Roadmap
52177 hepatocyte-ENCODE,HepG2-Roadmap,large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
52193 hepatocyte-ENCODE
52215 large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
52238 large_intestine_fetal-Roadmap,small_intestine_fetal-Roadmap
52283 liver-ENCODE
52345 HepG2-Roadmap,liver-ENCODE
68172 liver-ENCODE
68173 hepatocyte-ENCODE
68174 HepG2-Roadmap
68175 hepatocyte-ENCODE,HepG2-Roadmap,liver-ENCODE
tissues.uniq
7686 liver
7781 liver
7797 liver
7798 liver
7799 liver
7801 liver
7815 liver
7831 liver
7846 liver
7867 liver
7894 liver
30929 liver
30935 liver
30939 intestine,liver
30946 intestine
31061 intestine
46386 liver
46391 intestine
46392 intestine
46393 liver
46394 intestine
46395 intestine
46398 intestine
46488 liver
51946 liver
52046 intestine
52075 intestine
52123 intestine,liver
52149 liver
52156 intestine
52160 liver
52164 liver
52166 intestine
52168 liver
52171 intestine,liver
52177 intestine,liver
52193 liver
52215 intestine
52238 intestine
52283 liver
52345 liver
68172 liver
68173 liver
68174 liver
68175 liver
[ reached 'max' / getOption("max.print") -- omitted 92 rows ]
options(max.print = 75)
List of the infered new genes¶
options(max.print = 1000)
genes_dist1
[1] PDE4DIP LOC100996724 SEC22B NOTCH2NL
[5] NBPF10 LINC01719 HFE2 TXNIP
[9] POLR3GL ANKRD34A LIX1L RBM8A
[13] GNRHR2 PEX11B ANKRD35 PIAS3
[17] NUDT17 RNF115 POLR3C PDZK1
[21] GPR89A PRKAB2 PDIA3P1 FMO5
[25] CHD1L ITGA10 BCL9 DIP2B
[29] ATF1 METTL7A SLC11A2 TMPRSS12
[33] LETMD1 CSRNP2 TFCP2 POU6F1
[37] DAZAP2 SMAGP BIN2 GALNT6
[41] SLC4A8 ANKRD33 ACVRL1 ACVR1B
[45] GRASP NR4A1 ATG101 KRT80
[49] KRT7 KRT18 KRT8 EIF4B
[53] TNS2 LOC283335 SPRYD3 IGFBP6
[57] SOAT2 CSAD ZNF740 ITGB7
[61] RARG MFSD5 ESPL1 PFDN5
[65] C12orf10 AAAS AMHR2 PRR13
[69] PCBP2 MAP3K12 TARBP2 ATF7
[73] ATP5G2 CALCOCO1 CISTR HOXC10
[77] HOXC-AS3 HOXC-AS1 HOXC9 HOXC8
[81] HOXC4 HOXC6 HOXC5 LOC100240735
[85] SMUG1 CBX5 HNRNPA1 HNRNPA1P10
[89] NFE2 COPZ1 LOC102724050 ITGA5
[93] NCKAP1L PDE1B PPP1R1A HIGD2B
[97] BBS4 ADPGK-AS1 ADPGK NEO1
[101] NPTN CD276 LOXL1 LOXL1-AS1
[105] STOML1 PML ISLR STRA6
[109] CCDC33 SEMA7A UBL7 UBL7-AS1
[113] ARID3B EDC3 CYP1A1 CSK
[117] ULK3 SCAMP2 MPI FAM219B
[121] COX5A RPP25 SCAMP5 PPCDC
[125] C15orf39 COMMD4 NEIL1 MAN2C1
[129] PTPN9 CSPG4 ODF3L1 UBE2Q2
[133] FBXO22 TMEM266 CRNDE IRX5
[137] MMP2 LPCAT2 CES1 LOC283856
[141] GNAO1 DKFZP434H168 AMFR NUDT21
[145] OGFOD1 BBS2 MT2A MT1L
[149] MT1E MT1M MT1A MT1DP
[153] MT1B MT1F MT1G MT1H
[157] MT1X NUP93 HERPUD1 CPNE2
[161] FAM192A RSPRY1 ARL2BP PLLP
[165] CX3CL1 COQ9 CIAPIN1 POLR2C
[169] DOK4 CCDC102A ADGRG1 ADGRG3
[173] DRC7 KATNB1 CSNK2A2 CCDC113
[177] GINS3 SETD6 CES1P1 KIFC3
[181] ZNF319 USB1 SLC38A7 GOT2
[185] LOC388282 MMP15 CFAP20 NDRG4
[189] ADGRG5 CNOT1 FXYD5 LSR
[193] USF2 HAMP MAG LINC01531
[197] FFAR2 WTIP ZNF792 HPN
[201] LGI4 FXYD1 TMEM147 TMEM147-AS1
[205] HAUS5 RBM42 ETV2 COX6B1
[209] UPK1A-AS1 KMT2B IGFLR1 U2AF1L4
[213] PSENEN LIN37 HSPB6 PROSER3
[217] ARHGAP33 PRODH2 APLP1 NFKBID
[221] HCST LRFN3 SDHAF1 ALKBH6
[225] LOC101927572 BBS5 KLHL41 FASTKD1
[229] CCDC173 PHOSPHO2-KLHL23 PHOSPHO2 LINC01124
[233] SP5 ERICH2 LOC101926913 GAD1
[237] CYBRD1 GORASP2 TLK1 METTL8
[241] DCAF17 DYNC1I2 SLC25A12 HAT1
[245] METAP1D DLX1 DLX2 DLX2-AS1
[249] ITGA6 PDK1 CDCA7 RAPGEF4-AS1
[253] RAPGEF4 ZAK LINC01960 OLA1
[257] CIR1 SCRN3 GPR155 CHN1
[261] GULP1 COL3A1 COL5A2 WDR75
[265] SLC40A1 ASNSD1 ANKAR OSGEPL1-AS1
[269] OSGEPL1 PMS1 ORMDL1 C2orf88
[273] HIBCH MFSD6 NEMP2 INPP1
[277] NAB1 STAT4 MYO1B GLS
[281] STAT1 LOC105373805 NABP1 LOC105747689
[285] SDPR TMEFF2 FOXRED2 EIF3D
[289] IFT27 NCF4 CSF2RB TST
[293] MPST KCTD17 TMPRSS6 IL2RB
[297] C1QTNF6 SSTR3 RAC2 CYTH4
[301] SDHAP1 TFRC SLC51A LINC00885
[305] ZDHHC19 TCTEX1D2 TM4SF19-AS1 UBXN7-AS1
[309] UBXN7 RNF168 LY86 RREB1
[313] SSR1 CAGE1 RIOK1 SNRNP48
[317] BMP6 BLOC1S5 BLOC1S5-TXNDC5 EEF1E1
[321] EEF1E1-BLOC1S5 LOC101927972 DSP TXNDC5
[325] SLC35B3 LOC100506207 HULC NRSN1
[329] KAAG1 DCDC2 MRS2 GPLD1
[333] ALDH5A1 KIAA0319 TDP2 ACOT13
[337] C6orf62 GMNN C6orf229 RIPOR2
[341] CMAHP CARMIL1 SCGN HIST1H2AA
[345] HIST1H2APS1 SLC17A4 SLC17A1 TRIM38
[349] HIST1H3A HIST1H4A HIST1H4B HIST1H3B
[353] HIST1H2AB HIST1H1C HFE HIST1H4C
[357] HIST1H2BC HIST1H2AC HIST1H1D HIST1H4F
[361] HIST1H3F HIST1H2BH HIST1H3G HIST1H2BI
[365] HIST1H4H BTN3A2 BTN2A2 BTN3A1
[369] BTN2A3P BTN3A3 BTN2A1 ABT1
[373] ZNF322 HIST1H1A HIST1H2BB HIST1H3C
[377] LOC105374988 LOC100270746 LOC108783645 HIST1H1T
[381] HIST1H1E HIST1H2BD HIST1H2BE HIST1H4D
[385] HIST1H2AD HIST1H3D HIST1H2BF HIST1H4E
[389] ZSCAN25 LAMTOR4 C7orf43 GPC2
[393] STAG3 GATS PILRA PPP1R35
[397] TSC22D4 NYAP1 AGFG2 SAP25
[401] ZASP LRCH4 FBXO24 PCOLCE
[405] PCOLCE-AS1 MOSPD3 TFR2 GNB2
[409] POP7 EPO EPHB4 SLC12A9
[413] TRIP6 SRRT MUC3A LOC102724094
[417] VGF LOC101927746 GIGYF1 ACTL6B
[421] LOC105375429 UFSP1 DOK2 XPO7
[425] NPM2 DMTN FAM160B2 NUDT18
[429] HR REEP4 BMP1 PHYHIP
[433] POLR3D SLC39A14 PPP3CC SORBS3
[437] PDLIM2 C8orf58 CCAR2 BIN3-IT1
[441] BIN3 RHOBTB2 LOC286059 TNFRSF10B
[445] TNFRSF10D TNFRSF10A LOC389641 R3HCC1
[449] LOC100507156 LOXL2 ENTPD4 SLC25A37
[453] EGR3 LOC254896 ADAM28 ADAMDEC1
[457] PEBP4
17527 Levels: A1BG A1BG-AS1 A1CF A2M A2M-AS1 A3GALT2 A4GALT AAAS AACS ... ZZZ3
length(genes_dist1)
[1] 457
options(max.print = 75)