Original ABC model predictions on K562¶
Adapted from ABC model's README on github.
The Activity-by-contact model predicts which enhancers regulate which genes on a cell type specific basis.
Characteristics for this run¶
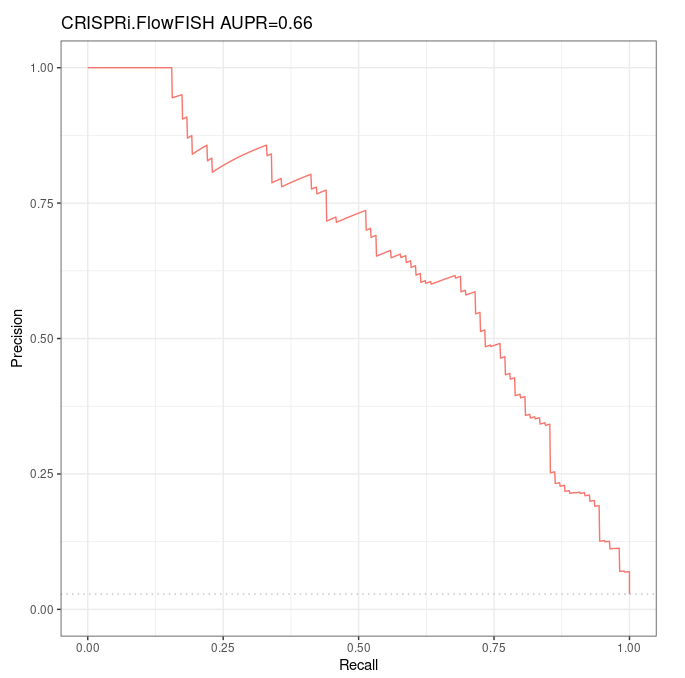
- Here we verify that we find the same AUPR as Fulco et al, based on their (109 ground positives, 3754 ground negatives) CRISPRi-FlowFISH dataset, available in
/work2/project/regenet/results/gscreen/homo_sapiens/hg19/crispr-flowfish/k562(private) - These tables already contain the ABC scores, so there should be no difference at all between the AUPR and Precision-Recall curves we compute and the ones in Fulco et al paper. The objective was only to ensure we used the expected data (later on we shall verify that we obtain the same ABC scores if we re-run the ABC model starting with these data).
Preprocessing¶
To obtain the \texttt{CRiFF} reference set we first downloaded Table S6a from Fulco et al 2019 as a tsv file, and then obtained the 109 positive and the 3754 negative E/G relationships performing the following filters (the negatives are indeed defined as either not significant or not associated to a decrease in gene expression):
awk 'NR==2{gsub(/\ /,".",$0); header=$0; OFS="\t";
print header > "109.fulco.positives.tsv";
print header > "3754.fulco.negatives.tsv"}
NR>=3{
split($0,a,"\t");
if(a[6]!="promoter"&&(a[10]=="TRUE"||a[17]>0.8))
{
if(a[10]=="TRUE"&&a[8]<0)
{
print > "109.fulco.positives.tsv";
}
else
{
print > "3754.fulco.negatives.tsv";
}}}' tableS6a.tsv
Merging:
awk 'BEGIN{FS="\t"; OFS="\t"} {if(NR==1){printf "interaction\t"} else{printf "1\t"}; print $1, $2, $3, $5, $19, $22, $23}' 109.fulco.positives.tsv > 3863.fulco.tsv
tail -n +2 3754.fulco.negatives.tsv |awk 'BEGIN{FS="\t"; OFS="\t"} {print 0, $1, $2, $3, $5, $19, $22, $23}' >> 3863.fulco.tsv
$ wc -l 3863.fulco.tsv 3864
Precision-Recall curves¶
We obtain the very same Precision-Recall curve and AUPR as Fulco et al (which was totally expected as we used their data directly, without computing anything by ourselves).