Guidelines to run the Average rank method over BENGI for GM12878¶
Prerequisites¶
Step 1: run Distance method¶
See dedicated guide.
Step 2: run DNase-expression correlation method¶
See dedicated guide.
Partial reimplementation of Run-Average-Rank.sh¶
We replaced the content of local_Scripts/Unsupervised-Methods/Run-Average-Rank.sh with the following:
#!/bin/bash
data=$1
version=$2
# Inserm computer
#workDir=~/Documents
# Personal computer
#workDir=~/Documents/INSERM
# Genotoul
workDir=/work2/project/regenet/workspace/thoellinger
distance=$workDir/BENGI/Distance-Method/Results/$data-Distance.$version.txt
correlation=$workDir/BENGI/Correlation-Methods/Sheffield/Results/$data-Correlation.$version.txt
output_dir=$workDir/BENGI/Correlation-Methods/Average-Rank
mkdir -p $output_dir
# for old format of distance method resulting files (with 2 columns),
# with "." as local decimal separator in bash, use:
#paste $distance $correlation | awk '{print $1 "\t" $2 "\t" $4}' | \
# sort -k2,2gr | awk 'BEGIN{x=0;r=0}{if (x != $2) r +=1; print $0 \
# "\t" r; x=$2}' | sort -k3,3gr | awk 'BEGIN{x=0;r=0}{if (x != $3) \
# r +=1; print $0 "\t" r "\t" ($NF+r)/2; x=$3}' | sort -k6,6g | \
# awk '{print $1 "\t" 1/$6 "\t" $2 "\t" $3 "\t" $4 "\t" $5 "\t" $6}' > $output_dir/$data-Average-Rank.$version.txt
# for new format of distance method resulting files (with 4 columns), use:
# if the local decimal separator in bash is "," (personal computer, genotoul, ...)
paste $distance $correlation |awk '{print $1 "\t" $2 "\t" $6}' |tr . , | \
sort -k2,2gr | awk 'BEGIN{x=0;r=0}{if (x != $2) r +=1; print $0 \
"\t" r; x=$2}' | sort -k3,3gr | awk 'BEGIN{x=0;r=0}{if (x != $3) \
r +=1; print $0 "\t" r "\t" ($NF+r)/2; x=$3}' | sort -k6,6g | \
awk '{print $1 "\t" 1/$6 "\t" $2 "\t" $3 "\t" $4 "\t" $5 "\t" $6}' |tr , . > $output_dir/$data-Average-Rank.$version.txt
# if the local decimal separator in bash is ".":
#paste $distance $correlation | awk '{print $1 "\t" $2 "\t" $6}' | \
# sort -k2,2gr | awk 'BEGIN{x=0;r=0}{if (x != $2) r +=1; print $0 \
# "\t" r; x=$2}' | sort -k3,3gr | awk 'BEGIN{x=0;r=0}{if (x != $3) \
# r +=1; print $0 "\t" r "\t" ($NF+r)/2; x=$3}' | sort -k6,6g | \
# awk '{print $1 "\t" 1/$6 "\t" $2 "\t" $3 "\t" $4 "\t" $5 "\t" $6}' > $output_dir/$data-Average-Rank.$version.txt
Note that we replaced
sort -k3,3gr | awk 'BEGIN{x=0;r=0}{if (x != $2) \ r +=1; print $0 "\t" r "\t" ($NF+r)/2; x=$2}'
which seems to be an error, with
sort -k3,3gr | awk 'BEGIN{x=0;r=0}{if (x != $3) \ r +=1; print $0 "\t" r "\t" ($NF+r)/2; x=$3}'
The reason is that the code does as follows:
- first, distance and correlation results (which are assumed sorted the very same way - which is the case if they were computed using the dedicated guides mentioned in the prerequisites) are merged
- then, the ranks of inverse distances are computed (the smaller the distance, the smaller the rank) with
sort -k2,2gr | awk 'BEGIN{x=0;r=0}{if (x != $2) r +=1; print $0 "\t" r; x=$2}'. So the condition onx=$2($2being the inverse distance) ensures the rank remains the same when consecutive inverse distances are constant. - then, the rank of the DNase-expression correlation method, plus the average distance-correlation rank, are computed (the greater the correlation, the smaller the rank) with
sort -k3,3gr | awk 'BEGIN{x=0;r=0}{if (x != $3) r +=1; print $0 "\t" r "\t" ($NF+r)/2; x=$3}'. So a condition on$2(value of the inverse distance) instead of$3(the correlation) would not make sense.
In the analysis, we will see that using $2 as Moore et al, leads to the very same results (AUPR of predictions over the 6 BENGI for GM12878) as in their paper ; and using $3, which we think is the correct way to do ; gives similar but slightly better results!
One should also note that we added tr . , at the beginning of the code and tr , . at the end. This is needed on every computer where bash default decimal separator is "," instead of ".".
At the end of the day, the fields are as follows:
- interaction (ground truth)
- final score = 1 / (average rank), such that the smaller the rank, the greater the score (=> high score = interaction suspected)
- inverse distance
- DNase-expression correlation
- inverse distance rank (the smaller the distance, the smaller the rank)
- DNase-expression correlation rank (the greater the correlation, the smaller the rank)
- average rank (= 1 over final score)
Running the code for BENGI benchmarks over GM12878¶
If working on Genotoul:
conda activate base && conda activate abcmodel && conda activate py2 && module load bioinfo/bedtools-2.27.1
srun --pty bash
Now:
./local_Scripts/Unsupervised-Methods/Run-Average-Rank.sh GM12878.CHiC v3
./local_Scripts/Unsupervised-Methods/Run-Average-Rank.sh GM12878.CTCF-ChIAPET v3
etc
Analysis with R¶
Code¶
.badCode {
background-color: #C9DDE4;
}
library(knitr)
## Global options
options(max.print="75")
opts_chunk$set(echo=TRUE,
cache=FALSE,
prompt=FALSE,
tidy=TRUE,
comment=NA,
message=FALSE,
warning=FALSE,
class.source="badCode")
opts_knit$set(width=75)
library(ggplot2)
library(ggpubr) # for ggarrange
library(dplyr) # for bind_rows
# Tools for precision-recall : (see https://classeval.wordpress.com/tools-for-roc-and-precision-recall/)
library(precrec)
#library(ROCR)
#library(pROC)
#library(PRROC)
rm(list = ls())
# Personal
work_dir = "~/Documents/INSERM/"
# Inserm
#work_dir = "~/Documents/"
path_to_results = paste(work_dir, "BENGI/Correlation-Methods/Average-Rank/", sep='')
file_names = c("GM12878.CHiC-Average-Rank.v3.txt", "GM12878.CTCF-ChIAPET-Average-Rank.v3.txt", "GM12878.GEUVADIS-Average-Rank.v3.txt", "GM12878.GTEx-Average-Rank.v3.txt", "GM12878.HiC-Average-Rank.v3.txt", "GM12878.RNAPII-ChIAPET-Average-Rank.v3.txt")
short_names = c('CHiC', 'CTCF', 'GEUVAVDIS', 'GTEx', 'HiC', 'RNAPII')
nb_files = length(file_names)
colnames <- c('interaction', 'score', 'inverse.distance', 'correlation', 'inverse.distance.rank', 'correlation.rank', 'average.rank')
results <- sapply(file_names, simplify=FALSE, function(file_name){
Df <- read.table(paste(path_to_results, as.character(file_name), sep=''), sep='\t')
Df[[1]] <- factor(Df[[1]], levels=c(0,1), labels=c("no interaction", "interaction"))
names(Df) <- colnames
return(Df)
})
names(results) <- short_names
#library(dplyr)
All_results <- bind_rows(results, .id = 'method') # requires library(dplyr)
str(results)
summary(results$CHiC)
ggplot(aes(y = average.rank, x = method, fill = interaction), data = All_results) + geom_boxplot()
sscurves_avg_rank <- list()s
sscurves_avg_rank <- sapply(results, simplify=FALSE, function(Df){
evalmod(scores = Df$score, labels = Df$interaction) # comes with "precrec" library
})
#library(ggplot2)
p1 <- autoplot(sscurves_avg_rank[[1]], curvetype = c("PRC")) + ggtitle(paste(short_names[1], signif(attr(sscurves_avg_rank[[1]], 'auc')[[4]][2], digits=2), sep = " AUPR="))
p2 <- autoplot(sscurves_avg_rank[[2]], curvetype = c("PRC")) + ggtitle(paste(short_names[2], signif(attr(sscurves_avg_rank[[2]], 'auc')[[4]][2], digits=2), sep = " AUPR="))
p3 <- autoplot(sscurves_avg_rank[[3]], curvetype = c("PRC")) + ggtitle(paste(short_names[3], signif(attr(sscurves_avg_rank[[3]], 'auc')[[4]][2], digits=2), sep = " A="))
p4 <- autoplot(sscurves_avg_rank[[4]], curvetype = c("PRC")) + ggtitle(paste(short_names[4], signif(attr(sscurves_avg_rank[[4]], 'auc')[[4]][2], digits=2), sep = " AUPR="))
p5 <- autoplot(sscurves_avg_rank[[5]], curvetype = c("PRC")) + ggtitle(paste(short_names[5], signif(attr(sscurves_avg_rank[[5]], 'auc')[[4]][2], digits=2), sep = " AUPR="))
p6 <- autoplot(sscurves_avg_rank[[6]], curvetype = c("PRC")) + ggtitle(paste(short_names[6], signif(attr(sscurves_avg_rank[[6]], 'auc')[[4]][2], digits=2), sep = " AUPR="))
# ggarrange comes with library('ggpubr')
figure <- ggarrange(p1, p2, p3, p4, p5, p6,
ncol = 3, nrow = 2)
figure
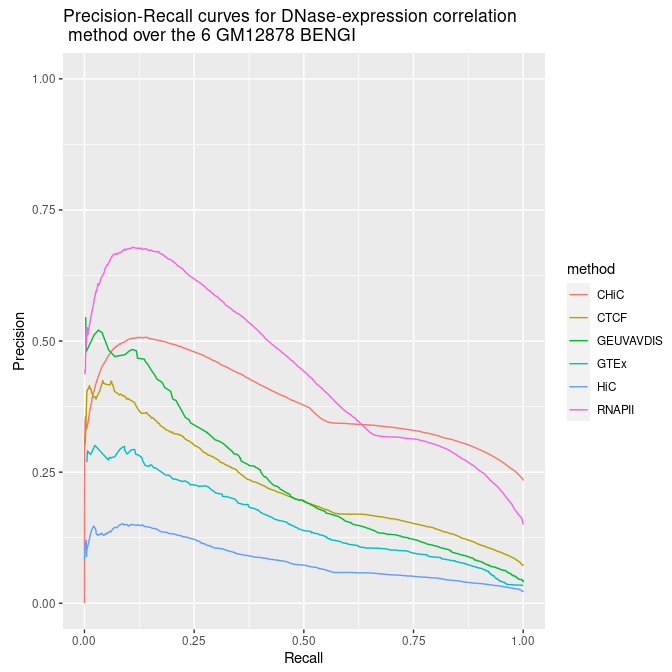
Alternatively we can plots all Precision - Recall curves on the same graph.
all_sscurves_avg_rank <- sapply(sscurves_avg_rank, simplify = FALSE, function(obj){
Df <- data.frame(obj$prcs[[1]]$x, obj$prcs[[1]]$y)
names(Df) <- c('x', 'y')
return(Df)
})
all_sscurves_avg_rank <- bind_rows(all_sscurves_avg_rank, .id = 'method')
total_nrow = nrow(all_sscurves_avg_rank)
max_nb_points_to_plot = 20000
if(total_nrow>max_nb_points_to_plot){
set.seed(1)
samples_points = sample(1:total_nrow, min(max_nb_points_to_plot, total_nrow), replace=TRUE)
} else{
samples_points = 1:total_nrow
}
ggplot(aes(y = y, x = x, color = method), data = all_sscurves_avg_rank[samples_points,]) + geom_line() + ylim(0,1) + xlab('Recall') + ylab('Precision') + ggtitle("Precision-Recall curves for DNase-expression correlation\n method over the 6 GM12878 BENGI")
Results¶
One can also directly have a look at the summarized results here
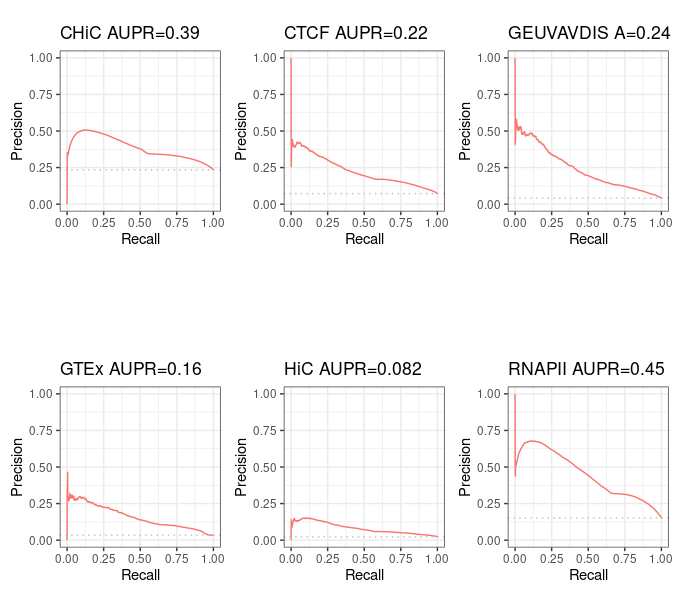
Not correcting the error in Run-Average-Rank.sh gives same results as in the paper¶
If we keep the small mistake found in Run-Average-Rank.sh, we find (hopefully!) the very same Precision - Recall curves as authors:
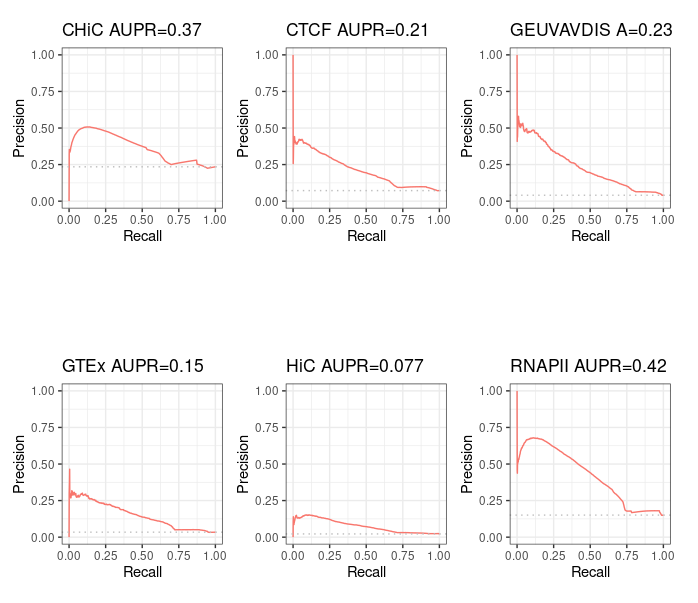
Correct results¶
If we correct the small error found in Run-Average-Rank.sh, we obtain the following results.